Table of Contents
Fits a series of peaks using 3D profile fitting as an Ikeda-Carpenter function by a bivariate gaussian.
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| OutputPeaksWorkspace | Output | Workspace | Mandatory | PeaksWorkspace with integrated peaks |
| OutputParamsWorkspace | Output | Workspace | Mandatory | MatrixWorkspace with fit parameters |
| InputWorkspace | Input | Workspace | Mandatory | An input Sample MDHistoWorkspace or MDEventWorkspace in HKL. |
| PeaksWorkspace | Input | Workspace | Mandatory | PeaksWorkspace with peaks to be integrated. |
| RunNumber | Input | number | 0 | Run Number to integrate |
| DQPixel | Input | number | 0.003 | The side length of each voxel in the non-MD histogram used for fitting (1/Angstrom) |
| UBFile | Input | string | File containing the UB Matrix in ISAW format. Allowed extensions: [‘.mat’] | |
| ModeratorCoefficientsFile | Input | string | File containing the Pade coefficients describing moderator emission versus energy. Allowed extensions: [‘.dat’] | |
| StrongPeakParamsFile | Input | string | File containing strong peaks profiles. If left blank, no profiles will be enforced. Allowed extensions: [‘.pkl’] | |
| IntensityCutoff | Input | number | 0 | Minimum number of counts to force a profile |
| EdgeCutoff | Input | number | 0 | Pixels within EdgeCutoff from a detector edge will be have a profile forced. Currently for 256x256 cameras only. |
| FracHKL | Input | number | 0.5 | Fraction of HKL to consider for profile fitting. |
| FracStop | Input | number | 0.05 | Fraction of max counts to include in peak selection. |
| MinpplFrac | Input | number | 0.9 | Min fraction of predicted background level to check |
| MaxpplFrac | Input | number | 1.1 | Max fraction of predicted background level to check |
| MindtBinWidth | Input | number | 15 | Smallest spacing (in microseconds) between data points for TOF profile fitting. |
| NTheta | Input | number | 50 | Number of bins for bivarite Gaussian along the scattering angle. |
| NPhi | Input | number | 50 | Number of bins for bivariate Gaussian along the azimuthal angle. |
| DQMax | Input | number | 0.15 | Largest total side length (in Angstrom) to consider for profile fitting. |
| DtSpread | Input | number | 0.03 | The fraction of the peak TOF to consider for TOF profile fitting. |
| PeakNumber | Input | number | -1 | Which Peak to fit. Leave negative for all. |
This algorithm performs integration of single crystal Bragg peaks by fitting the intensity distribution as a 3D distribution made of an Ikeda-Carpenter function (TOF coordinate) and a bivariate Normal distribution.
See IntegratePeaksMD v2 or IntegrateEllipsoids v1 for peak-minus-background integration algorithms in reciprocal space.
The algorithms takes two input workspaces:
This algorithm will fit a set of peaks in a PeaksWorkspace. The intensity profile is fit to an MDHisto workspace formed around the peak location.
To construct the measured distribution to be fit, a histogram of events is made around the peak. This histogram is in (qx, qy, qz) and composed of voxels of side length dQPixel. To minimize the effect of neighboring peaks on profile fitting, the variable qMask is used to only consider a region around the peak in (h,k,l) space. It will filter voxels outside of (h ± fracHKL, k ± fracHKL, l ± fracHKL) from calculations used for profile fitting. In practice, values of 0.35 < fracHKL < 0.5 seem to work best.
The time-of-flight (TOF), t, of each voxel is determined as:
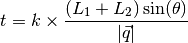
The events are histogrammed by their t values to create a TOF profile. This profile can then be fit to the Ikeda Carpenter function. To separate the peak and background, different levels of intensity are filtered out. The predicted background level is determined as the average background not near the peak or off the edge and values within minppl_frac and maxppl_frac times the predicted value are tried. The best fit to the expected moderator emission (determined by the moderator coefficients defined in ModeratorCoefficientsFile) is taken and these voxels are considered to be signal.
TOF goes as 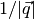 and so it is natural to use spherical coordinate. In that sense, the other two coordinates
are
and so it is natural to use spherical coordinate. In that sense, the other two coordinates
are 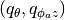 - along the scattering angle and azimuthal angle, respectively. From the
MDHisto Workspace (filtered by qMask and using only the signal voxels from the TOF fight), a 2D histogram is constructed
which is fit to a bivariate normal distribution. The histogram has nTheta
- along the scattering angle and azimuthal angle, respectively. From the
MDHisto Workspace (filtered by qMask and using only the signal voxels from the TOF fight), a 2D histogram is constructed
which is fit to a bivariate normal distribution. The histogram has nTheta  nPhi bins.
nPhi bins.
For weak peaks or peaks near detector edges, the 2D histogram likely does not reflect the full profile. To address this, the
profile of the nearest strong peaks is forced when doing the BVG fit. The profile is fit (allowed to vary 10% in
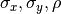 ) and location and amplitude are not fixed. Weak peaks are defined as peaks with fewer
than forceCutoff counts from peak-minus-background integration or within dEdge pixels of the detector edge.
) and location and amplitude are not fixed. Weak peaks are defined as peaks with fewer
than forceCutoff counts from peak-minus-background integration or within dEdge pixels of the detector edge.
The final intensity profile is given by
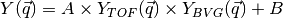
where 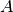 and
and 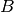 are scaling constants. Here the background is assumed to be constant
are scaling constants. Here the background is assumed to be constant 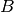 over the volume of
the peak, so the model of the peak itself is
over the volume of
the peak, so the model of the peak itself is 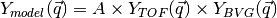 .
The peak intensity
.
The peak intensity 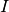 , is given by summing
, is given by summing  over voxels which are greater than FracStop of the maximum.
over voxels which are greater than FracStop of the maximum.
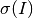 is given as
is given as
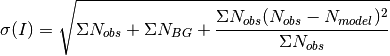
where the first two terms come from Poissionian statistics and the final term is the variance of the fit. Those sums are over the same voxels used to calculate intensity.
Example - IntegratePeaksProfileFitting
1 2 3 4 5 6 7 8 9 10 11 12 13 14 | Load(Filename='/SNS/MANDI/IPTS-8776/0/5921/NeXus/MANDI_5921_event.nxs', OutputWorkspace='MANDI_5921_event')
MANDI_5921_md = ConvertToMD(InputWorkspace='MANDI_5921_event', QDimensions='Q3D', dEAnalysisMode='Elastic',
Q3DFrames='Q_lab', QConversionScales='Q in A^-1',
MinValues='-5, -5, -5', Maxvalues='5, 5, 5', MaxRecursionDepth=10,
LorentzCorrection=False)
LoadIsawPeaks(Filename='/SNS/MANDI/shared/ProfileFitting/demo_5921.integrate', OutputWorkspace='peaks_ws')
IntegratePeaksProfileFitting(OutputPeaksWorkspace='peaks_ws_out', OutputParamsWorkspace='params_ws',
InputWorkspace='MANDI_5921_md', PeaksWorkspace='peaks_ws', RunNumber=5921, DtSpread=0.015,
UBFile='/SNS/MANDI/shared/ProfileFitting/demo_5921.mat',
ModeratorCoefficientsFile='/SNS/MANDI/shared/ProfileFitting/franz_coefficients_2017.dat',
MinpplFrac=0.9, MaxpplFrac=1.1, MindtBinWidth=15,
StrongPeakParamsFile='/SNS/MANDI/shared/ProfileFitting/strongPeakParams_beta_lac_mut_mbvg.pkl',
peakNumber=30)
|
Categories: Algorithms | Crystal\Integration
Python: IntegratePeaksProfileFitting.py (last modified: 2018-06-22)