\(\renewcommand\AA{\unicode{x212B}}\)
PDConvertRealSpace v1¶
Summary¶
Transforms a Workspace2D between different real space functions.
See Also¶
Properties¶
Name |
Direction |
Type |
Default |
Description |
|---|---|---|---|---|
InputWorkspace |
Input |
Mandatory |
Input workspace. The units are assumed to be distance |
|
From |
Input |
string |
G(r) |
Function type in the input workspace. Allowed values: [‘G(r)’, ‘GK(r)’, ‘g(r)’] |
To |
Input |
string |
G(r) |
Function type in the output workspace. Allowed values: [‘G(r)’, ‘GK(r)’, ‘g(r)’] |
OutputWorkspace |
Output |
Mandatory |
Output workspace |
Description¶
The neutron diffraction is measuring the differential scattering cross section. This can be converted to the structure factor \(S(Q)\). Using the PDFFourierTransform algorithm, one can obtain the pair distribution function, \(G(r)\):
One can transform between this quantity and \(GK(r)\) or \(g(r)\) using:
where \(\rho_0\) is the sample number density and \(\langle b_{coh} \rangle^2\) is defined in the Materials concept page.
NOTE: This algorithm requires that SetSampleMaterial v1 is called prior in order to determine the \(\rho_0\) and \(\langle b_{coh} \rangle^2\) terms.
PyStoG¶
This algorithm uses the external project PyStoG and specifically uses the pystog.converter.Converter object. To modify the underlying algorithms, the following functions are used for the conversions.
- \(G(r)\) conversions are:
To \(G_K(r)\) see
pystog.converter.Converter.G_to_GK()To \(g(r)\) see
pystog.converter.Converter.G_to_g()
- \(G_K(r)\) conversions are:
To \(G(r)\) see
pystog.converter.Converter.GK_to_G()To \(g(r)\) see
pystog.converter.Converter.GK_to_g()
- \(g(r)\) conversions are:
To \(G(r)\) see
pystog.converter.Converter.g_to_G()To \(GK(r)\) see
pystog.converter.Converter.g_to_GK()
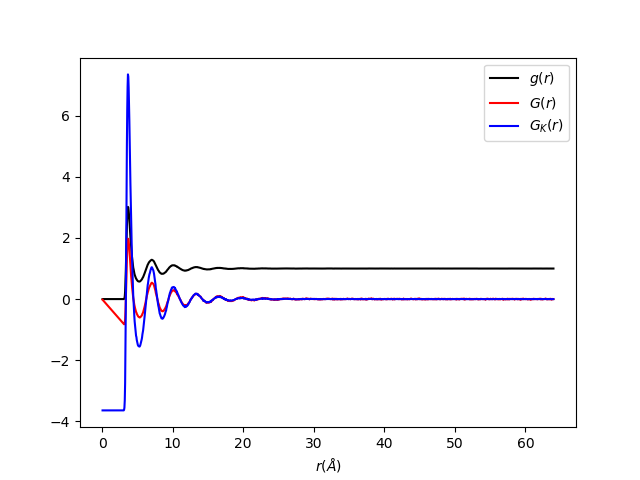
Usage¶
import wget
import numpy as np
import matplotlib.pyplot as plt
from mantid.simpleapi import CreateWorkspace
# Grab the real data for argon
url = "https://raw.githubusercontent.com/marshallmcdonnell/pystog/master/tests/test_data/argon.real_space.dat"
filename = wget.download(url)
r, gofr, GofR_, GKofR_ = np.loadtxt(filename, skiprows=2, unpack=True)
# Convert gofr to Mantid wksp
g_of_r = CreateWorkspace(DataX=r, DataY=gofr,
UnitX="Angstrom",
Distribution=True)
SetSampleMaterial(InputWorkspace=g_of_r, ChemicalFormula='Ar')
bigG_of_r=PDConvertRealSpace(InputWorkspace=g_of_r, From='g(r)', To='G(r)')
GK_of_r=PDConvertRealSpace(InputWorkspace=g_of_r, From='g(r)', To='GK(r)')
fig, ax = plt.subplots(subplot_kw={'projection':'mantid'})
ax.plot(g_of_r,'k-', label='$g(r)$')
ax.plot(bigG_of_r,'r-', label='$G(r)$')
ax.plot(GK_of_r,'b-', label='$G_K(r)$')
ax.legend() # show the legend
ax.set_xlabel(r'$r(\AA)$')
fig.show()
The output should look like:
Categories: AlgorithmIndex | Diffraction\Utility
Source¶
Python: PDConvertRealSpace.py