SANSILLAutoProcess v1¶
Summary¶
Performs complete SANS data reduction at the ILL.
See Also¶
Properties¶
Name |
Direction |
Type |
Default |
Description |
|---|---|---|---|---|
OutputWorkspace |
Output |
WorkspaceGroup |
Mandatory |
The output workspace group containing reduced data. |
SampleRuns |
Input |
list of str lists |
Sample run(s). Allowed values: [‘nxs’] |
|
AbsorberRuns |
Input |
list of str lists |
Absorber (Cd/B4C) run(s). Allowed values: [‘nxs’] |
|
BeamRuns |
Input |
list of str lists |
Empty beam run(s). Allowed values: [‘nxs’] |
|
FluxRuns |
Input |
list of str lists |
Empty beam run(s) for flux calculation only; if left blank flux will be calculated from BeamRuns. Allowed values: [‘nxs’] |
|
ContainerRuns |
Input |
list of str lists |
Empty container run(s). Allowed values: [‘nxs’] |
|
SampleTransmissionRuns |
Input |
list of str lists |
Sample transmission run(s). Allowed values: [‘nxs’] |
|
ContainerTransmissionRuns |
Input |
list of str lists |
Container transmission run(s). Allowed values: [‘nxs’] |
|
TransmissionBeamRuns |
Input |
list of str lists |
Empty beam run(s) for transmission. Allowed values: [‘nxs’] |
|
TransmissionAbsorberRuns |
Input |
list of str lists |
Absorber (Cd/B4C) run(s) for transmission. Allowed values: [‘nxs’] |
|
ThetaDependent |
Input |
boolean |
True |
Whether or not to use 2theta dependent transmission correction |
SensitivityMaps |
Input |
string |
File(s) or workspaces containing the maps of relative detector efficiencies. |
|
DefaultMaskFile |
Input |
string |
File or workspace containing the default mask (typically the detector edges and dead pixels/tubes) to be applied to all the detector configurations. |
|
MaskFiles |
Input |
string |
File(s) or workspaces containing the detector mask (typically beam stop). |
|
ReferenceFiles |
Input |
string |
File(s) or workspaces containing the corrected water data (in 2D) for absolute normalisation. |
|
SolventFiles |
Input |
string |
File(s) or workspaces containing the corrected solvent data (in 2D) for solvent subtraction. |
|
SensitivityOutputWorkspace |
Output |
The output sensitivity map workspace. |
||
NormaliseBy |
Input |
string |
Timer |
Choose the normalisation type. Allowed values: [‘None’, ‘Timer’, ‘Monitor’] |
SampleThickness |
Input |
number |
0.1 |
Sample thickness [cm] |
TransmissionBeamRadius |
Input |
number |
0.1 |
Beam radius [m]; used for transmission calculations. |
BeamRadius |
Input |
dbl list |
0.1 |
Beam radius [m]; used for beam center finding and flux calculations. |
WaterCrossSection |
Input |
number |
1 |
Provide water cross-section; used only if the absolute scale is done by dividing to water. |
MaxQxy |
Input |
dbl list |
-1 |
Maximum of absolute Qx and Qy. |
DeltaQ |
Input |
dbl list |
-1 |
The dimension of a Qx-Qy cell. |
OutputPanels |
Input |
boolean |
False |
Whether or not process the individual detector panels. |
OutputType |
Input |
string |
I(Q) |
Choose the output type. Allowed values: [‘I(Q)’, ‘I(Qx,Qy)’, ‘I(Phi,Q)’] |
CalculateResolution |
Input |
string |
None |
Choose to calculate the Q resolution. Allowed values: [‘MildnerCarpenter’, ‘DirectBeam’, ‘None’] |
DefaultQBinning |
Input |
string |
PixelSizeBased |
Choose how to calculate the default Q binning. Allowed values: [‘PixelSizeBased’, ‘ResolutionBased’] |
BinningFactor |
Input |
number |
1 |
Specify a multiplicative factor for default Q binning (pixel or resolution based). |
NPixelDivision |
Input |
number |
1 |
Number of subpixels to split the pixel (NxN) |
NumberOfWedges |
Input |
number |
0 |
Number of wedges to integrate separately. |
WedgeAngle |
Input |
number |
30 |
Wedge opening angle [degrees]. |
WedgeOffset |
Input |
number |
0 |
Wedge offset angle from x+ axis. |
AsymmetricWedges |
Input |
boolean |
False |
Whether to have asymmetric wedges. |
IQxQyLogBinning |
Input |
boolean |
False |
I(Qx, Qy) log binning when binning is not specified. |
WavelengthRange |
Input |
dbl list |
1,10 |
Wavelength range [Angstrom] to be used in integration (TOF only). |
OutputBinning |
Input |
string |
Output binning for each distance. : separated list of binning params. |
|
ClearCorrected2DWorkspace |
Input |
boolean |
True |
Whether to clear the fully corrected 2D workspace. |
SensitivityWithOffsets |
Input |
boolean |
False |
Whether the sensitivity data has been measured with different horizontal offsets. |
StitchReferenceIndex |
Input |
number |
1 |
Index of reference workspace during stitching. |
ShapeTable |
Input |
The name of the table workspace containing drawn shapes on which to integrate. If provided, NumberOfWedges, WedgeOffset and WedgeAngle arguments are ignored. |
||
Wavelength |
Input |
number |
0 |
Wavelength set for the data, will override nexus, intended for D16 reduction. |
MinThreshold |
Input |
number |
0 |
Minimum threshold for calculated efficiency. |
MaxThreshold |
Input |
number |
2 |
Maximum threshold for calculated efficiency. |
Description¶
This algorithms performs complete treatment of SANS data recorded with the ILL instruments D11, D22 and D33. This high level algorithm steers the reduction and performs the full set of corrections for a given sample run; measured with one or more detector distances.
The sample measurement will be corrected for all the effects and converted to Q-space, producing by default the azimuthal average curve \(I(Q)\). One can use water reference measurement in order to derive the relative inter-pixel sensitivity map of the detector or the reduced water data for absolute normalisation of the subsequent samples. The regular output will contain fully corrected water run, and there will be an additional output containing the sensitivity map itself. The sensitivity map, as well as reduced water can be saved out to a file and used for sample reductions.
OutputBinning¶
This property can be used to set manual q-binning parameters per detector distance/configuration. This accepts a string, which must be : delimited sets of binning parameters per distance. The reason it is a string, is that at a given configuration the parameters set can be composed of , separated values (such as start, width, end). In fact, it could be even more complicated. See the semantics in SANSILLIntegration. If one parameter set is provided, i.e. there is no :, the same parameter set will be used for all the detector distances/configurations.
Caching with ADS¶
This algorithm does not clean-up the intermediate workspaces after execution. This is done intentionally for performance reasons. For example, once the transmission of a sample is calculated, it will be reused for further iterations of processing of the same sample at different detector distances. As other example, once the container is processed at a certain distance, it will be reused for all the subsequent samples measured at the same distance, if the container run is the same. The same caching is done for absorber, empty beam, container, sensitivity and mask workspaces. The caching relies on Analysis Data Service (ADS) through naming convention by appending the relevant process name to the run number. When multiple runs are summed, the run number of the first run is attributed to the summed workspace name.
Note
To run these usage examples please first download the usage data, and add these to your path. In Mantid this is done using Manage User Directories.
Example - full treatment of 3 samples at 3 different distances in D11
beams = '2866,2867+2868,2878'
containers = '2888+2971,2884+2960,2880+2949'
container_tr = '2870+2954'
beam_tr = '2867+2868'
samples = ['2889,2885,2881',
'2887,2883,2879',
'3187,3177,3167']
sample_tr = ['2871', '2869', '3172']
thick = [0.1, 0.2, 0.2]
# reduce samples
for i in range(len(samples)):
SANSILLAutoProcess(
SampleRuns=samples[i],
BeamRuns=beams,
ContainerRuns=containers,
MaskFiles='mask1.nxs,mask2.nxs,mask3.nxs',
SensitivityMaps='sens-lamp.nxs',
SampleTransmissionRuns=sample_tr[i],
ContainerTransmissionRuns=container_tr,
TransmissionBeamRuns=beam_tr,
SampleThickness=thick[i],
OutputWorkspace='iq_s' + str(i + 1)
)
print('Distance 1 Q-range:{0:4f}-{1:4f} AA'.format(mtd['iq_s1_#1_d39.0m_c40.5m_w5.6A'].readX(0)[0],
mtd['iq_s1_#1_d39.0m_c40.5m_w5.6A'].readX(0)[-1]))
print('Distance 2 Q-range:{0:4f}-{1:4f} AA'.format(mtd['iq_s1_#2_d8.0m_c8.0m_w5.6A'].readX(0)[0],
mtd['iq_s1_#2_d8.0m_c8.0m_w5.6A'].readX(0)[-1]))
print('Distance 3 Q-range:{0:4f}-{1:4f} AA'.format(mtd['iq_s1_#3_d2.0m_c5.5m_w5.6A'].readX(0)[0],
mtd['iq_s1_#3_d2.0m_c5.5m_w5.6A'].readX(0)[-1]))
Output:
Distance 1 Q-range:0.001350-0.020028 AA
Distance 2 Q-range:0.007625-0.092061 AA
Distance 3 Q-range:0.032571-0.343635 AA
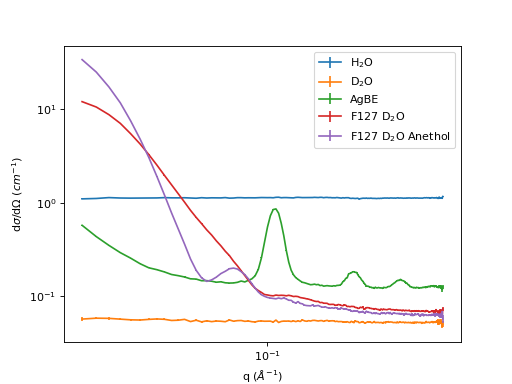
Example 2 - full treatment of 5 samples in D33
from mantid.simpleapi import *
import matplotlib.pyplot as plt
config.setFacility('ILL')
config['default.instrument'] = 'D33'
config.appendDataSearchSubDir('ILL/D33/')
absorber = '002227'
tr_beam = '002192'
can_tr = '002193'
empty_beam = '002219'
can = '002228'
mask = 'D33Mask2.nxs'
sample_names = ['H2O', 'D2O', 'AgBE', 'F127_D2O', 'F127_D2O_Anethol']
sample_legends = ['H$_2$O', 'D$_2$O', 'AgBE', 'F127 D$_2$O', 'F127 D$_2$O Anethol']
samples = ['002229', '001462', '001461', '001463', '001464']
transmissions = ['002194', '002195', '', '002196', '002197']
# Autoprocess every sample
for i in range(len(samples)):
SANSILLAutoProcess(
SampleRuns=samples[i],
SampleTransmissionRuns=transmissions[i],
MaskFiles=mask,
AbsorberRuns=absorber,
BeamRuns=empty_beam,
ContainerRuns=can,
ContainerTransmissionRuns=can_tr,
TransmissionBeamRuns=tr_beam,
CalculateResolution='None',
OutputWorkspace=sample_names[i]
)
fig, ax = plt.subplots(subplot_kw={'projection':'mantid'})
plt.yscale('log')
plt.xscale('log')
# Plot the result of every autoprocess
for wName in sample_names:
ax.errorbar(mtd[wName][0])
plt.legend(sample_legends)
ax.set_ylabel('d$\sigma$/d$\Omega$ ($cm^{-1}$)')
#fig.show()
(Source code, png, hires.png, pdf)
Categories: AlgorithmIndex | ILL\SANS | ILL\Auto
Source¶
Python: SANSILLAutoProcess.py