SavePlot1D v1¶
Summary¶
Save 1D plots to a file
See Also¶
Properties¶
Name |
Direction |
Type |
Default |
Description |
|---|---|---|---|---|
InputWorkspace |
Input |
Mandatory |
Workspace to plot |
|
OutputFilename |
Input |
string |
Name of the image file to savefile. Allowed extensions: [‘.png’] |
|
OutputType |
Input |
string |
image |
Method for rendering plot. Allowed values: [‘image’] |
XLabel |
Input |
string |
Label on the X axis. If empty, it will be taken from workspace |
|
YLabel |
Input |
string |
Label on the Y axis. If empty, it will be taken from workspace |
|
SpectraList |
Input |
int list |
Which spectra to plot |
|
SpectraNames |
Input |
str list |
Override with custom names for spectra |
|
Result |
Output |
string |
||
PopCanvas |
Input |
boolean |
False |
If true, a Matplotlib canvas will be popped out, which contains the saved plot. |
Description¶
Save 1D plots to a png file, as part of autoreduction. Multiple spectra in the same workspace will be represented by curves on the same plot. Grouped workspaces will be shown as subplots. If the workspace has more than one spectra, but less or equal to ten, labels will be shown.
Note
The figures contain lines between points, no error bars.
Note
Requires matplotlib version>= 1.2.0
Usage¶
#create some workspaces
CreateWorkspace(OutputWorkspace='w1',DataX='1,2,3,4,5,1,2,3,4,5',DataY='1,4,5,3,2,2,3,1',DataE='1,2,2,1,1,1,1,1',NSpec='2',UnitX='DeltaE')
CreateWorkspace(OutputWorkspace='w2',DataX='1,2,3,4,5,1,2,3,4,5',DataY='4,2,5,3,3,1,3,1', DataE='1,2,2,1,1,1,1,1',NSpec='2',UnitX='Momentum',VerticalAxisUnit='Wavelength',VerticalAxisValues='2,3',YUnitLabel='Something')
wGroup=GroupWorkspaces("w1,w2")
#write to a file
try:
import mantid
filename=mantid.config.getString("defaultsave.directory")+"SavePlot1D.png"
SavePlot1D(InputWorkspace=wGroup,OutputFilename=filename)
except:
pass
The file should look like
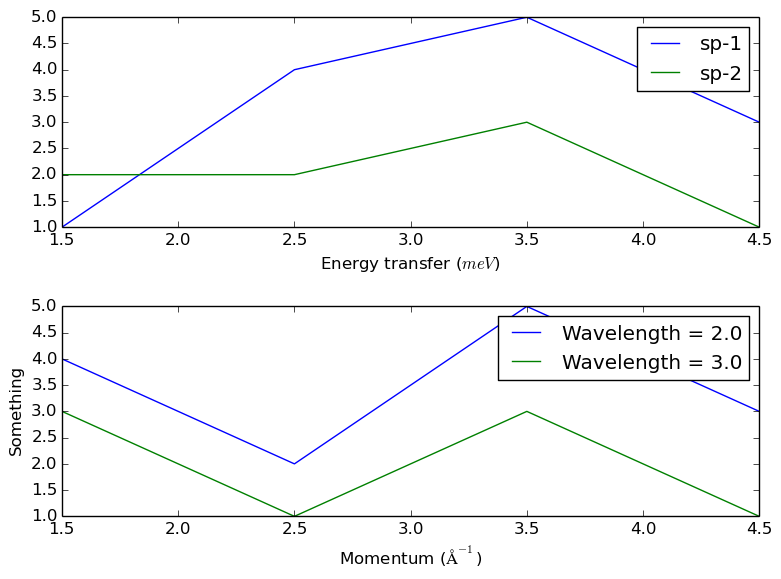
Usage in autoreduction¶
# create some workspaces
CreateWorkspace(OutputWorkspace='w1',DataX='1,2,3,4,5,1,2,3,4,5',DataY='1,4,5,3,2,2,3,1',DataE='1,2,2,1,1,1,1,1',NSpec='2',UnitX='DeltaE')
# get the div html/javascript for the plot
div = SavePlot1D(InputWorkspace='w1', OutputType='plotly')
from postprocessing.publish_plot import publish_plot
request = publish_plot('TESTINSTRUMENT', 12345, files={'file':div})
print("post returned {}".format(request.status_code))
To see what the result looks like on your local system, add the
Filename argument (.html extension) and change to
OutputType='plotly-full'.
Categories: AlgorithmIndex | DataHandling\Plots
Source¶
Python: SavePlot1D.py