Table of Contents
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| InputWorkspace | Input | IMDWorkspace | Mandatory | An input Sample MDHistoWorkspace or MDEventWorkspace in HKL. |
| DeltaHKL | Input | number | 0.5 | Distance from integer HKL to integrate peak. |
| GridPoints | Input | number | 201 | Number of grid points for each dimension of HKL box. |
| NeighborPoints | Input | number | 10 | Number of points in 5^3 surrounding points above intensity threshold for point to be part of peak. |
| FluxWorkspace | Input | MatrixWorkspace | An optional input workspace containing momentum dependent flux for normalization. | |
| SolidAngleWorkspace | Input | MatrixWorkspace | An optional input workspace containing momentum integrated vanadium for normalization (a measure of the solid angle). | |
| PeaksWorkspace | Input | PeaksWorkspace | Mandatory | A PeaksWorkspace containing the peaks to integrate. |
| OutputWorkspace | Output | PeaksWorkspace | The output PeaksWorkspace will be a copy of the input PeaksWorkspace with the peaks’ integrated intensities. |
IntegratePeaksMDHKL provides integration of a MDHistoWorkspace or MDEventWorkspace in 3-dimensions. The units of the workspace must be HKL. The main usage will be for data normalized by MDNormSCD. A 3D box is created for each peak and the background and peak data are separated. The intensity and sigma of the intensity is found from the grid inside the peak and the background is subtracted. The boxes are created and integrated in parallel and less memory is required than binning all HKL at once.
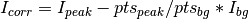
with the errors summed in quadrature:
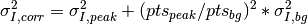
Using the DeltaHKL parameter, the problem of nearly peaks or regions of diffuse scattering can be avoided. Also for normalized data, the unmeasured data points are excluded from the background. See white regions in last figure.
Example - IntegratePeaksMDHKL event histo
#Create PeaksWorkspace
sampleWs = CreateSampleWorkspace()
pws = CreatePeaksWorkspace(InstrumentWorkspace=sampleWs,NumberOfPeaks=3)
p = pws.getPeak(0)
p.setHKL(5,0,0)
p = pws.getPeak(1)
p.setHKL(0,0,0)
p = pws.getPeak(2)
p.setHKL(-5,0,0)
#Test with MDEventWorkspace
mdws = CreateMDWorkspace(Dimensions=3, Extents=[-10,10,-10,10,-10,10], Names='[H,0,0],[0,K,0],[0,0,L]',Units='A^-1,A^-1,A^-1',Frames='HKL,HKL,HKL')
FakeMDEventData(InputWorkspace=mdws, PeakParams=[100000,-5,0,0,1])
FakeMDEventData(InputWorkspace=mdws, PeakParams=[100000,0,0,0,1])
FakeMDEventData(InputWorkspace=mdws, PeakParams=[100000,5,0,0,1])
pws =IntegratePeaksMDHKL(InputWorkspace=mdws,PeaksWorkspace=pws,DeltaHKL=1.5,GridPoints=21)
for i in range(3):
p = pws.getPeak(i)
print p.getIntensity(),p.getSigmaIntensity()
#Test with MDHistoWorkspace
mdws = BinMD(InputWorkspace=mdws,AlignedDim0="[H,0,0],-10,10,101",AlignedDim1="[0,K,0],-10,10,101",AlignedDim2="[0,0,L],-10,10,101")
pws =IntegratePeaksMDHKL(InputWorkspace=mdws,PeaksWorkspace=pws,DeltaHKL=1.5,GridPoints=21)
for i in range(3):
p = pws.getPeak(i)
print p.getIntensity(),p.getSigmaIntensity()
Output:
99913.3212993 316.123352485
99913.3212993 316.123352485
99913.3212993 316.123352485
99945.2374619 316.157736186
99951.6716018 316.164910575
99926.3456269 316.133414893
Categories: Algorithms | MDAlgorithms\Peaks