Table of contents
There are six workflow algorithms supporting data reduction at ILL’s time-of-flight instruments. These algorithms are:
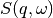 .
.The algorithms can be used as flexible building blocks in Python scripts. Not all of them need to be necessarily used in a reduction: the simplest script could call DirectILLCollectData v1 and DirectILLReduction v1 only.
Together with the other algorithms and services provided by the Mantid framework, the reduction algorithms can handle a great number of reduction scenarios. If this proves insufficient, however, the algorithms can be accessed using Python. Before making modifications it is recommended to copy the source files and rename the algorithms as not to break the original behavior.
This document tries to give an overview on how the algorithms work together via Python examples. Please refer to the algorithm documentation for details of each individual algorithm.
A very basic reduction would include a vanadium reference and a sample and follow the steps:
These steps would translate to something like the following simple Python script:
# Add a temporary data search directory.
mantid.config.appendDataSearchDir('/data/')
# Vanadium
DirectILLCollectData(
Run='0100:0109',
OutputWorkspace='vanadium',
OutputEPPWorkspace='vanadium-epps', # Elastic peak positions.
OutputRawWorkspace='vanadium-raw' # 'Raw' data for diagnostics.
)
DirectILLIntegrateVanadium(
InputWorkspace='vanadium',
OutputWorkspace='integrated',
EPPWorkspace='vanadium-epps'
)
DirectILLDiagnostics(
InputWorkspace='vanadium-raw',
OutputWorkspace='diagnostics',
EPPWorkspace='vanadium-epps',
RawWorkspace='vanadium-raw'
)
# Sample
DirectILLCollectData(
Run='0201, 0205, 0209-0210',
OutputWorkspace='sample'
)
DirectILLReduction(
InputWorkspace='sample',
OutputWorkspace='SofQW',
IntegratedVanadiumWorkspace='integrated'
DiagnosticsWorkspace='diagnostics'
)
Every DirectILL algorithm has an OutputWorkspace property which provides the main output workspace. Additionally, the algorithms may provide optional output workspaces to be used with other algorithms or for reporting/debugging purposes. The linking of outputs to inputs is an important feature of the DirectILL algorithms and allows for great flexibility in the reduction. An example of the usage of these optional output workspaces is the OutputEPPWorkspace which in the vanadium case above is needed in the integration and diagnostics steps:
...
# Vanadium
DirectILLCollectData(
...
OutputEPPWorkspace='vanadium-epps' # This workspace...
)
DirectILLIntegrateVanadium(
...
EPPWorkspace='vanadium-epps' # ...is needed here...
)
DirectILLDiagnostics(
...
EPPWorkspace='vanadium-epps' # ...and here.
)
...
As shown above, these optional outputs are sometimes named similarly the corresponding inputs giving a hint were they are supposed to be used.
A more complete reduction example would include corrections for self-shielding:
The above workflow would translate to this kind of Python script:
# Add a temporary data search directory.
mantid.config.appendDataSearchDir('/data/')
# Vanadium
DirectILLCollectData(
Run='0100:0109',
OutputWorkspace='vanadium',
OutputEPPWorkspace='vanadium-epps', # Elastic peak positions.
OutputRawWorkspace='vanadium-raw' # 'Raw' data for diagnostics.
)
DirectILLIntegrateVanadium(
InputWorkspace='vanadium',
OutputWorkspace='integrated',
EPPWorkspace='vanadium-epps'
)
DirectILLDiagnostics(
InputWorkspace='vanadium-raw',
OutputWorkspace='diagnostics',
EPPWorkspace='vanadium-epps',
RawWorkspace='vanadium-raw'
)
# Sample
DirectILLCollectData(
Run='0201, 0205, 0209-0210',
OutputWorkspace='sample',
)
geometry = {
'Shape': 'FlatPlate',
'Width': 4.0,
'Height': 5.0,
'Thick': 0.1,
'Center': [0.0, 0.0, 0.0],
'Angle': 45.0
}
material = {
'ChemicalFormula': 'Ni Cr Fe',
'SampleNumberDensity': 0.09
}
SetSample(
InputWorkspace='sample',
Geometry=geometry,
Material=material
)
DirectILLSelfShielding(
InputWorkspace='sample',
OutputWorkspace='corrections'
)
DirectILLApplySelfShielding(
InputWorkspace='sample',
OutputWorkspace='sample-corrected',
SelfShieldingCorrectionWorkspace='corrections',
)
DirectILLReduction(
InputWorkspace='sample-corrected',
OutputWorkspace='SofQW',
IntegratedVanadiumWorkspace='integrated'
DiagnosticsWorkspace='diagnostics'
)
Mantid can be picky with binning when doing arithmetics between workspaces. This is an issue for the time-of-flight instruments at ILL as the time axis needs to be corrected to correspond to a physical flight distance. Even thought data is recorded with the same nominal wavelength, the actual value written in the NeXus files may differ between runs. Incident energy calibration further complicates matters. As the correction to the time-of-flight axis depends on the wavelength, two datasets loaded into Mantid with DirectILLCollectData v1 may have slightly different time-of-flight axis. This prevents arithmetics between the workspaces. The situation is most often encountered between sample and the corresponding empty container.
To alleviate the situation, the output workspaces of DirectILLCollectData v1 can be forced to use the same wavelength. The following Python script shows how to propagate the calibrated incident energy from the first loaded workspace into the rest:
DirectILLCollectData(
Run='0100:0109',
OutputWorkspace='sample1',
OutputIncidentEnergyWorkspace='Ei' # Get a common incident energy.
)
# Empty container.
DirectILLCollectData(
Run='0201:0205',
OutputWorkspace='container',
IncidentEnergyWorkspace='Ei' # Empty container should have same TOF binning.
)
# More samples with same nominal wavelength and container as 'sample1'.
runs = ['0110:0119,', '0253:0260']
index = 1
for run in runs:
DirectILLCollectData(
Run=run,
OutputWorkspace='sample{}'.format(index),
IncidentEnergyWorkspace='Ei'
)
index += 1
# The empty container is now compatible with all the samples.
The container background subtraction is done perhaps a bit counterintuitively in DirectILLApplySelfShielding v1. At the moment the self-shielding corrections and the empty container data do not have much to do with each other but this may change in the future if the so called Paalman-Pings corrections are used.
With empty container data, the steps to reduce the experimental data might look like this:
A corresponding Python script follows.
mantid.config.appendDataSearchDir('/data/')
# Vanadium
DirectILLCollectData(
Run='0100:0109',
OutputWorkspace='vanadium',
OutputEPPWorkspace='vanadium-epps',
OutputRawWorkspace='vanadium-raw'
)
DirectILLIntegrateVanadium(
InputWorkspace='vanadium',
OutputWorkspace='integrated',
EPPWorkspace='vanadium-epps'
)
DirectILLDiagnostics(
InputWorkspace='vanadium-raw',
OutputWorkspace='diagnostics',
EPPWorkspace='vanadium-epps',
RawWorkspace='vanadium-raw'
)
# Sample
DirectILLCollectData(
Run='0201, 0205, 0209-0210',
OutputWorkspace='sample',
OutputIncidentEnergyWorkspace='Ei'
)
# Container
DirectILLCollectData(
Run='0333:0335',
OutputWorkspace='container',
IncidentEnergyWorkspace='Ei'
)
# Container self-shielding.
# Geometry XML allows for very complex geometries.
containerShape = """
<hollow-cylinder id="inner-ring">
<centre-of-bottom-base x="0.0" y="-0.04" z="0.0" />
<axis x="0.0" y="1.0" z="0.0" />
<inner-radius val="0.017" />
<outer-radius val="0.018" />
<height val="0.08" />
</hollow-cylinder>
<hollow-cylinder id="outer-ring">
<centre-of-bottom-base x="0.0" y="-0.04" z="0.0" />
<axis x="0.0" y="1.0" z="0.0" />
<inner-radius val="0.02" />
<outer-radius val="0.021" />
<height val="0.08" />
</hollow-cylinder>
<algebra val="inner-ring : outer-ring" />
"""
geometry = {
'Shape': 'CSG',
'Value': containerShape
}
material = {
'ChemicalFormula': 'Al',
'SampleNumberDensity': 0.09
}
SetSample(
InputWorkspace='container',
Geometry=geometry,
Material=material
)
DirectILLSelfShielding(
InputWorkspace='container',
OutputWorkspace='container-corrections'
)
DirectILLApplySelfShielding(
InputWorkspace='container',
OutputWorkspace='container-corrected',
SelfShieldingCorrectionWorkspace='container-corrections',
)
# Sample self-shielding and container subtraction.
geometry = {
'Shape': 'HollowCylinder',
'Height': 8.0,
'InnerRadius': 1.8,
'OuterRadium': 2.0,
'Center': [0.0, 0.0, 0.0]
}
material = {
'ChemicalFormula': 'C2 O D6',
'SampleNumberDensity': 0.1
}
SetSample('sample', geometry, material)
DirectILLSelfShielding(
InputWorkspace='sample',
OutputWorkspace='sample-corrections'
)
DirectILLApplySelfShielding(
InputWorkspace='sample',
OutputWorkspace='sample-corrected',
SelfShieldingCorrectionWorkspace='sample-corrections',
EmptyContainerWorkspace='container-corrected'
)
DirectILLReduction(
InputWorkspace='sample-corrected',
OutputWorkspace='SofQW',
IntegratedVanadiumWorkspace='integrated'
DiagnosticsWorkspace='diagnostics'
)
Sometimes the empty container is not measured at all the experiment’s temperature points. One can use Mantid’s workspace arithmetics to perform simple linear interpolation in temperature:
# Container measurement temperatures.
T0 = 3.0
T1 = 250.0
DT = T1 - T0
# Target sample temperature.
Ts = 190.0
# Linear interpolation.
container_190 = (T1 - Ts) / DT * mtd['container_3'] + (Ts - T0) / DT * mtd['container_250']
DirectILLApplySelfShielding(
InputWorkspace='sample',
EmptyContainerWorkspace=container_190
)
As usual, care should be taken when extrapolating the container data outside the measured range.
The reduction algorithms do not produce much output to Mantid logs by default. Also, none of the intermediate workspaces generated during the run of the DirectILL algorithms will show up in the analysis data service. Both behaviors can be controlled by the SubalgorithmLogging and Cleanup properties. Enabling SubalgorithmLogging will log all messages from child algorithms to Mantid’s logs. Disabling Cleanup will unhide the intermediate workspaces created during the algorithm run and disable their deletion.
Note, that disabling Cleanup might produce a large number of workspaces and cause the computer to run out of memory.
The intensities of individual pixels on IN5 are very low. This makes the fitting procedure employed by FindEPP v2 to work unreliably or fail altogether. Because of this, DirectILLCollectData v1 will use CreateEPP v1 instead by default for IN5. CreateEPP v1 produces an artificial EPP workspace based on the instrument geometry. This should be accurate enough for vanadium integration and diagnostics.
The elastic peak diagnostics might be usable to mask the beam stop of IN5. The background diagnostics, on the other hand, are turned off by default as it makes no sense to mask individual pixels based on them.
When working on memory constrained systems, it is recommended to manually delete the workspaces which are not needed anymore in the reduction script. The SaveNexus v1 can be used to save the data to disk.
Lets put it all together into a complex Python script. The script below reduces the following dataset:
mantid.config.appendDataSearchDir('/data/')
# Gather dataset information.
containerRuns = '96,97'
vanadiumRuns = '100-103'
# Samples at 50K, 100K and 150K.
# Wavelength 1
containerRuns1 = {
50: '131-137',
150: '138-143'
}
runs1 = {
50: '105, 107-110',
100: '112-117',
150: '119-123, 125'
}
# Wavelength 2
containerRun2 = '166-170'
runs2 = {
50: '146, 148, 150',
100: '151-156',
150: '160-165'
}
# Vanadium & vanadium container.
DirectILLCollectData(
Run=vanadiumRuns,
OutputWorkspace='vanadium',
OutputEPPWorkspace='vanadium-epp',
OutputRawWorkspace='vanadium-raw',
OutputIncidentEnergyWorkspace='vanadium-Ei' # Use for container
)
DirectILLCollectData(
Run=containerRuns,
OutputWorkspace='vanadium-container',
IncidentEnergyWorkspace='vanadium-Ei'
)
containerShape = """
<hollow-cylinder id="inner-ring">
<centre-of-bottom-base x="0.0" y="-0.04" z="0.0" />
<axis x="0.0" y="1.0" z="0.0" />
<inner-radius val="0.017" />
<outer-radius val="0.018" />
<height val="0.08" />
</hollow-cylinder>
<hollow-cylinder id="outer-ring">
<centre-of-bottom-base x="0.0" y="-0.04" z="0.0" />
<axis x="0.0" y="1.0" z="0.0" />
<inner-radius val="0.02" />
<outer-radius val="0.021" />
<height val="0.08" />
</hollow-cylinder>
<algebra val="inner-ring : outer-ring" />
"""
containerGeometry = {
'CSG': containerShape
}
containerMaterial = {
'ChemicalFormula': 'Al',
'SampleNumberDensity': 0.1
}
SetSample('vanadium-container', containerGeometry, containerMaterial)
DirectILLSelfShielding(
InputWorkspace='vanadium-container',
OutputWorkspace='vanadium-container-self-shielding'
)
DirectILLApplySelfShielding(
InputWorkspace='vanadium-container',
OutputWorkspace='vanadium-container-corrected'
SelfShieldingCorrectionWorkspace='vanadium-container-self-shielding'
)
sampleGeometry = {
'Shape': 'HollowCylinder',
'Height': 8.0,
'InnerRadius': 1.8,
'OuterRadium': 2.0,
'Center': [0.0, 0.0, 0.0]
}
vanadiumMaterial = {
'ChemicalFormula': 'V',
'SampleNumberDensity': 0.15
}
SetSample('vanadium', sampleGeometry, vanadiumMaterial)
DirectILLSelfShielding(
InputWorkspace='vanadium',
OutputWorkspace='vanadium-self-shielding'
)
DirectILLApplySelfShielding(
InputWorkspace='vanadium',
OutputWorkspace='vanadium-corrected',
SelfShieldingCorrectionWorkspace='vanadium-self-shielding',
EmptyContainerWorkspace='vanadium-container-corrected'
)
DirectILLIntegrateVanadium(
InputWorkspace='vanadium-corrected',
OutputWorkspace='vanadium-calibration',
EPPWorkspace='vanadium-epp'
)
diagnosticsResult = DirectILLDiagnoseDetectors(
InputWorkspace='vanadium-raw',
OutputWorkspace='mask',
EPPWorkspace='vanadium-epp',
OutputReportWorkspace='diagnostics-report'
)
# Sample and container at wavelength 1.
DirectILLCollectData(
Run=runs1[50],
OutputWorkspace='run1-50K',
OutputIncidentEnergyWorkspace='Ei1',
OutputFlatBkgWorkspace='bkg1-50K'
)
DirectILLCollectData(
Run=containerRuns1[50],
OutputWorkspace='container1-50K',
IncidentEnergyWorkspace='Ei1'
)
SetSample('container1-50K', containerGeometry, containerMaterial)
DirectILLSelfShielding(
InputWorkspace='container1-50K',
OutputWorkspace='container1-self-shielding'
)
DirectILLCollectData(
Run=containerRuns1[150],
OutputWorkspace='container1-150K',
IncidentEnergyWorkspace='Ei1'
)
interpolated = 0.5 * (mtd['container1-50K'] + mtd['container1-150K'])
RenameWorkspace(interpolated, 'container1-100K')
for T in [50, 100, 150]:
DirectILLApplySelfShielding(
InputWorkspace='container1-{}K'.format(T),
OutputWorkspace='container1-{}K-corrected'.format(T),
SelfShieldingCorrectionWorkspace='container1-self-shielding'
)
sampleMaterial = {
'ChemicalFormula': 'Fe 2 O 3',
'SampleNumberDensity': 0.23
}
SetSample('run1-50K', sampleGeometry, sampleMaterial)
DirectILLSelfShielding(
InputWorkspace='run1-50K',
OutputWorkspace='run1-self-shielding',
)
for T in runs1:
if T != 50:
# 50K data has been loaded already.
DirectILLCollectData(
Run=runs1[T],
OutputWorkspace='run1-{}K'.format(T),
IncidentEnergyWorkspace='Ei1',
FlatBkgWorkspace='bkg1-50K'
)
DirectILLApplySelfShielding(
InputWorkspace='run1-{}K'.format(T),
OutputWorkspace='run1-{}K-corrected'.format(T),
SelfShieldingCorrectionWorkspace='run1-self-shielding',
EmptyContainerWorkspace='container1-{}K-corrected'.format(T)
)
DirectILLReduction(
InputWorkspace='run1-{}K-corrected'.format(T),
OutputWorkspace='SofQW1-{}K'.format(T),
IntegratedVanadiumWorkspace='vanadium-calibration',
DiagnosticsWorkspace='mask'
)
SaveNexus('SofQW1-{}K'.format(T), '/data/output2-{}.nxs'.format(T))
# Sample and container at wavelength 2.
DirectILLCollectData(
Run=runs2[50],
OutputWorkspace='run2-50K',
OutputIncidentEnergyWorkspace=Ei2',
OutputFlatBkgWorkspace='bgk2-50K'
)
DirectILLCollectData(
Run=containerRun2,
OutputWorkspace='container2',
IncidentEnergyWorkspace='Ei2'
)
SetSample('container2', containerGeometry, containerMaterial)
DirectILLSelfShielding(
InputWorkspace='container2',
OutputWorkspace='container2-self-shielding'
)
DirectILLApplySelfShielding(
InputWorkspace='container2',
OutputWorkspace='container2-corrected',
SelfShieldingCorrectionWorkspace='container2-self-shielding'
)
SetSample('run2-50K', sampleGeometry, sampleMaterial)
DirectILLSelfShielding(
InputWorkspace='run2-50K',
OutputWorkspace='run2-self-shielding'
)
for T in runs2:
if T != 50:
# 50K data has been loaded already.
DirectILLCollectData(
Run=runs2[T]
OuputWorkspace='run2-{}K'.format(T),
IncidentEnergyWorkspace='Ei2',
FlatBkgWorkspace='bkg2-50K
)
DirectILLApplySelfShielding(
InputWorkspace='run2-{}K'.format(T),
OutputWorkspace='run2-{}K-corrected'.format(T),
SelfShieldingCorrectionWorkspace='run2-self-shielding',
EmptyContainerWorkspace='container2'
)
DirectILLReduction(
InputWorkspace='run2-{}K-corrected'.format(T),
OutputWorkspace='SofQW2-{}K'.format(T),
IntegratedVanadiumWorkspace='vanadium-calibration',
DiagnosticsWorkspace='mask'
)
SaveNexus('SofQW2-{}K'.format(T), '/data/output2-{}.nxs'.format(T))
Category: Techniques