Table of Contents
The program estimates the quasielastic components of each of the groups of spectra and requires the resolution file (.RES file) and optionally the normalisation file created by ResNorm.
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| InputType | Input | string | File | Origin of data input - File (.nxs) or Workspace. Allowed values: [‘File’, ‘Workspace’] |
| Instrument | Input | string | iris | Instrument. Allowed values: [‘irs’, ‘iris’, ‘osi’, ‘osiris’] |
| Analyser | Input | string | graphite002 | Analyser & reflection. Allowed values: [‘graphite002’, ‘graphite004’] |
| Program | Input | string | QL | Name of program to run. Allowed values: [‘QL’, ‘QSe’] |
| SamNumber | Input | string | Mandatory | Sample run number |
| ResInputType | Input | string | File | Origin of res input - File (_res.nxs) or Workspace. Allowed values: [‘File’, ‘Workspace’] |
| ResType | Input | string | Res | Format of Resolution file. Allowed values: [‘Res’, ‘Data’] |
| ResNumber | Input | string | Mandatory | Resolution run number |
| ResNorm | Input | boolean | False | Use ResNorm output file |
| ResNormInputType | Input | string | File | Origin of ResNorm input - File (_red.nxs) or Workspace. Allowed values: [‘File’, ‘Workspace’] |
| ResNormNumber | Input | string | ResNorm run number | |
| BackgroundOption | Input | string | Sloping | Form of background to fit. Allowed values: [‘Sloping’, ‘Flat’, ‘Zero’] |
| ElasticOption | Input | boolean | True | Include elastic peak in fit |
| FixWidth | Input | boolean | False | Fix one of the widths |
| WidthFile | Input | string | Name of file containing fixed width values | |
| EnergyMin | Input | number | -0.5 | Minimum energy for fit. Default=-0.5 |
| EnergyMax | Input | number | 0.5 | Maximum energy for fit. Default=0.5 |
| SamBinning | Input | number | 1 | Binning value (integer) for sample. Default=1 |
| ResBinning | Input | number | 1 | Binning value (integer) for resolution - QLd only. Default=1 |
| Sequence | Input | boolean | True | Switch Sequence Off/On |
| Plot | Input | string | None | Plot options. Allowed values: [‘None’, ‘ProbBeta’, ‘Intensity’, ‘FwHm’, ‘Fit’, ‘All’] |
| Verbose | Input | boolean | True | Switch Verbose Off/On |
| Save | Input | boolean | False | Switch Save result to nxs file Off/On |
The model that is being fitted is that of a δ-function (elastic
component) of amplitude 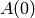 and Lorentzians of amplitude
and Lorentzians of amplitude
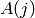 and HWHM
and HWHM 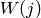 where
where 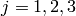 . The whole
function is then convolved with the resolution function. The -function
and Lorentzians are intrinsically normalised to unity so that the
amplitudes represent their integrated areas.
. The whole
function is then convolved with the resolution function. The -function
and Lorentzians are intrinsically normalised to unity so that the
amplitudes represent their integrated areas.
For a Lorentzian, the Fourier transform does the conversion:
![1/(x^{2}+\delta^{2}) \Leftrightarrow exp[-2\pi(\delta k)]](../_images/math/8cfe2d45d1b9de22f5e1b9f503b4a0353b75086f.png) . If
. If
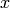 is identified with energy
is identified with energy 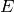 and
and 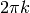 with
with
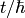 where t is time then:
where t is time then:
![1/[E^{2}+(\hbar / \tau )^{2}] \Leftrightarrow exp[-t /\tau]](../_images/math/286b48e50473ff75521e96f5cea3bcf14ce3f7c2.png) and
and
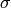 is identified with
is identified with 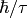 . The program
estimates the quasielastic components of each of the groups of spectra
and requires the resolution file and optionally the normalisation file
created by ResNorm.
. The program
estimates the quasielastic components of each of the groups of spectra
and requires the resolution file and optionally the normalisation file
created by ResNorm.
For a Stretched Exponential, the choice of several Lorentzians is
replaced with a single function with the shape :
![\psi\beta(x) \Leftrightarrow exp[-2\pi(\sigma k)\beta]](../_images/math/364a3414ec4a8c1e73d8b4a1f4bfed39ae740039.png) . This, in
the energy to time FT transformation, is
. This, in
the energy to time FT transformation, is
![\psi\beta(E) \Leftrightarrow exp[-(t/\tau)\beta]](../_images/math/a107edd8fafd9951836b41fc46f0bfdd89b363db.png) . So \sigma is
identified with
. So \sigma is
identified with 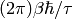 . The model that is fitted
is that of an elastic component and the stretched exponential and the
program gives the best estimate for the
. The model that is fitted
is that of an elastic component and the stretched exponential and the
program gives the best estimate for the 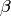 parameter and the
width for each group of spectra.
parameter and the
width for each group of spectra.
This routine was originally part of the MODES package.
Example - a basic example using QLines to fit a reduced workspace.
def createSampleWorkspace(name, random=False):
""" Creates a sample workspace with a single lorentzian that looks like IRIS data"""
import os
function = "name=Lorentzian,Amplitude=8,PeakCentre=5,FWHM=0.7"
ws = CreateSampleWorkspace("Histogram", Function="User Defined", UserDefinedFunction=function, XUnit="DeltaE", Random=True, XMin=0, XMax=10, BinWidth=0.01)
ws = CropWorkspace(ws, StartWorkspaceIndex=0, EndWorkspaceIndex=9)
ws = ScaleX(ws, -5, "Add")
ws = ScaleX(ws, 0.1, "Multiply")
#load instrument and instrument parameters
LoadInstrument(ws, InstrumentName='IRIS', RewriteSpectraMap=True)
path = os.path.join(config['instrumentDefinition.directory'], 'IRIS_graphite_002_Parameters.xml')
LoadParameterFile(ws, Filename=path)
ws = RenameWorkspace(ws, OutputWorkspace=name)
return ws
ws = createSampleWorkspace("irs26176_graphite002_red", random=True)
res = createSampleWorkspace("irs26173_graphite002_res")
QLines(SamNumber='26176', ResNumber='26173', InputType='Workspace', ResInputType='Workspace', Instrument='irs', Analyser='graphite002', Plot='None')
Categories: Algorithms | Workflow\MIDAS