Table of Contents
Panel parameters and L0 are optimized to minimize errors between theoretical and actual q values for the peaks
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| PeakWorkspace | InOut | PeaksWorkspace | Mandatory | Workspace of Indexed Peaks |
| a | Input | number | Optional | Lattice Parameter a (Leave empty to use lattice constants in peaks workspace) |
| b | Input | number | Optional | Lattice Parameter b (Leave empty to use lattice constants in peaks workspace) |
| c | Input | number | Optional | Lattice Parameter c (Leave empty to use lattice constants in peaks workspace) |
| alpha | Input | number | Optional | Lattice Parameter alpha in degrees (Leave empty to use lattice constants in peaks workspace) |
| beta | Input | number | Optional | Lattice Parameter beta in degrees (Leave empty to use lattice constants in peaks workspace) |
| gamma | Input | number | Optional | Lattice Parameter gamma in degrees (Leave empty to use lattice constants in peaks workspace) |
| ChangeL1 | Input | boolean | True | Change the L1(source to sample) distance |
| ChangePanelSize | Input | boolean | True | Change the height and width of the detectors. Implemented only for RectangularDetectors. |
| EdgePixels | Input | number | 0 | Remove peaks that are at pixels this close to edge. |
| DetCalFilename | Input | string | SCDCalibrate.DetCal | Path to an ISAW-style .detcal file to save. Allowed extensions: [‘.detcal’, ‘.det_cal’] |
| XmlFilename | Input | string | Path to an Mantid .xml description(for LoadParameterFile) file to save. Allowed extensions: [‘.xml’] | |
| ColFilename | Input | string | ColCalcvsTheor.nxs | Path to a NeXus file comparing calculated and theoretical column of each peak. Allowed extensions: [‘.nxs’] |
| RowFilename | Input | string | RowCalcvsTheor.nxs | Path to a NeXus file comparing calculated and theoretical row of each peak. Allowed extensions: [‘.nxs’] |
| TofFilename | Input | string | TofCalcvsTheor.nxs | Path to a NeXus file comparing calculated and theoretical TOF of each peak. Allowed extensions: [‘.nxs’] |
This algorithm calibrates panels of Rectangular Detectors
or packs of tubes in an instrument. The initial path,
panel centers and orientations are adjusted so the error in Q
positions from the theoretical Q positions is minimized.
Given a set of peaks indexed by 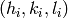 , we
modify the instrument parameters, p, and then find Q in the sample frame,
, we
modify the instrument parameters, p, and then find Q in the sample frame,
 that mininizes the following:
that mininizes the following:
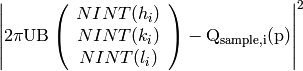
NINT is the nearest integer function. B is fixed from the input lattice parameters, but U is modified by CalculateUMatrix for all peaks before and after optimization. The initial path, L1, is optimized for all peaks before and after each panel or pack’s parameters are optimized. The panels and packs’ parameters are optimized in parallel. An option is available to adjust the panel widths and heights for Rectangular Detectors in a second iteration with all the other parameters fixed.
After calibration, you can save the workspace to Nexus (or Nexus processed) and get it back by loading in a later Mantid session. You can copy the calibration to another workspace using the same instrument by means of the CopyInstrumentParameters v1 algorithm. To do so select the workspace, which you have calibrated as the InputWorkspace and the workspace you want to copy the calibration to, the OutputWorkspace.
#Calibrate peaks file and load to workspace
LoadIsawPeaks(Filename='MANDI_801.peaks', OutputWorkspace='peaks')
#TimeOffset is not stored in xml file, so use DetCal output if you need TimeOffset
SCDCalibratePanels(PeakWorkspace='peaks',DetCalFilename='mandi_801.DetCal',XmlFilename='mandi_801.xml',a=74,b=74.5,c=99.9,alpha=90,beta=90,gamma=60)
LoadEmptyInstrument(Filename=config.getInstrumentDirectory() + 'MANDI_Definition_2013_08_01.xml', OutputWorkspace='MANDI_801_event_DetCal')
CloneWorkspace(InputWorkspace='MANDI_801_event_DetCal', OutputWorkspace='MANDI_801_event_xml')
LoadParameterFile(Workspace='MANDI_801_event_xml', Filename='mandi_801.xml')
LoadIsawDetCal(InputWorkspace='MANDI_801_event_DetCal', Filename='mandi_801.DetCal')
det1 = mtd['MANDI_801_event_DetCal'].getInstrument().getDetector(327680)
det2 = mtd['MANDI_801_event_xml'].getInstrument().getDetector(327680)
if det1.getPos() == det2.getPos():
print "matches"
Output:
matches
Categories: Algorithms | Crystal\Corrections