A wide variety of algorithms are available within Mantid for processing of data, writing results files, etc. These may be run interactively, one at a time, but in general scripting (using the Python scripting language) enables a series of algorithms to be called in a specific order to carry out more complex tasks, such as data normalisation and writing data files suitable for input to Rietveld refinement programmes (GSAS, FullProf, etc).
All the script files used to process GEM, HRPD, INS and Polaris data have been integrated to Mantid which can be found inside the following directory on a Windows machine: C:\MantidInstall\scripts\CryPowderISIS.
There are a preference file and a small script which is required to run inside Scripting Window on mantid in order to carry out the data normalisation:
The preference file, a file with an extension name of .pref (e.g: UserPrefFile.pref) will contain the following at least:
These directories have been left blank but can be changed and set by each user on their preference. The directories are also modifiable via small script which is ran inside Scripting Window
The script which is required to be written inside Scripting Window on Mantid contains:
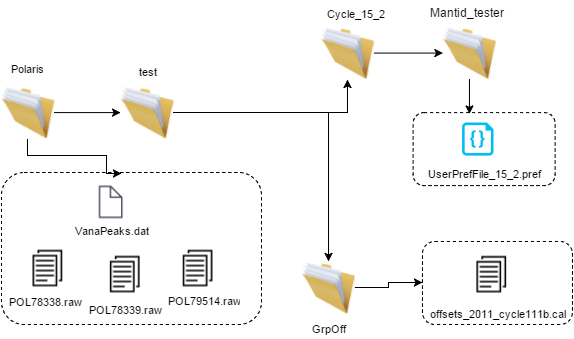
The vanadium and background files are smoothed (a spline wih 150 points works well with Polaris data), and the vanadium Bragg reflections removed. The high and low d-spacing limits of the observed V reflections in each of the detector banks are stored in a file called VanaPeaks.dat - which is stored inside Polaris folder in Files & Folder Post-Structure.
Both the smoothed and unsmoothed aligned and grouped vanadium data sets are written to the Polaris/test/Cycle_15_2/Calibration folder. These may be read in to a workspace and compared against one another to ensure that the smoothing and peak stripping is satisfactory (the smoothing function and number of ‘fixed’ points may be chosen to suit the data).
When ExistingV is yes inside the pref file that is being utilised, you are required to have nexus files named after the value of CorrVanFile which is also found inside the pref file. Within the image below we are using Polaris instrument so we shall need 5 nexus files as you can see inside the calibration folder (5 is the number of banks for Polaris). If the nexus files are not found, then the script will automatically change ExvistingV to no from yes until the end of the process.
Once the process has completed, files should be generated inside the Calibration and Mantid_tester folder.
The structure of the folder shown above in the image can be easily modified within the script that is written inside Scripting Window. The script that is being used to generate above folder structure can be found in the Usage section. If the Analysisdir equals empty string (“”) or Analysisdir is not pass as a parameter within the python script, the GrpOff and Cycle_15_2 folder will be required inside the Polaris folder.
Similarly if the Cycle_15_2 or user is passed as an empty string (“”) or else is not passed as a parameter, then the script shall look for the files in previous folder.
The ‘GrpOff’ folder is always required, within the ‘GrpOff’ folder is a .cal file which gives the detector grouping (i.e. the detector banks’ numbers) and the Offset to be applied to focus each detector (this is in addition to the “ideal” instrument geometry focusing information, which Mantid determines from the Instrument Definition File).
The run number, vanadium number and V-Empty number data is required to be provided within the raw directory, if the vana-peaks are also required to be removed then a file named VanaPeaks.dat is needed inside the raw directory. This will enable vana peaks to be removed by interpolation in range given within the VanaPeaks.dat file. The vanadium number and V-Empty number is set in the pref file as shown below, the script shall look for the raw files (with the format of <instrument><run-number>, e.g: POL78338 and POL78339) inside the raw directory.
#----------------------------------------------------
# Runs Numbers
#----------------------------------------------------
#
Vanadium 78338
V-Empty 78339
S-Empty 0
#
The output files will vary on the values provided in pref file for the following variables, which either equal yes or no.
#Output
XYE-TOF yes
XYE-D yes
GSS yes
Nexus yes
The XYE-TOF, XYE-D, GSS and Nexus files along with a copy of the Grouping file are all generated where the pref file is located, which would be inside the ‘Mantid_tester’ folder in Files & Folder Post-Structure.
The Calibration folder (which is created automatically), where that cycle’s smoothed and corrected vanadium files are stored (note - there is not a multiple scattering correction available yet for the vanadium). If a file name is not specified in the .pref file for the smoothed vanadium files, a file name is generated automatically, which contains both the vanadium and the background run numbers.
With the mantid feature Manage User Directories, users are able to reveal the instrument directory to mantid, which can then be utilised inside the python script by simply calling DIRS[0], if the following script is also passed in Scripting Window (DIR[0]- 0 being the first/top directory listed inside Manage User Directories).
from mantid import config
DIRS = config['datasearch.directories'].split(';')
To get the directory inside the Manage User Directories, just use Browse To Directory button the find the directory of the instrument folder, once directory has been added, select the directory and move it to the top of the list with the help of Move Up button on the right.
However depending on the preference of the user, a directory of the instrument can directly be passed as a variable, for example:
dir = 'X:\'
FilesDir = 'X:\Polaris'
expt = cry_ini.Files('Polaris', RawDir=FilesDir, Analysisdir='test', forceRootDirFromScripts=False, inputInstDir=dir)
User may also place the instrument folder where the script is located, which would be found in the following directory on Windows platform C:\MantidInstall\scriptsCryPowderISIS. Using instrument folder from where the scripts are located can simply be done by modifying the following line of the Usage script to:
expt = cry_ini.Files('Polaris', RawDir=(DIRS[0] + "Polaris"), Analysisdir='test', forceRootDirFromScripts=True)
Image below displays the only files required when ExistingV is no inside the pref file. This means that the Calibration folder, where that cycle’s smoothed and corrected vanadium files are stored will not be required for this process. Instead the files will be generated and the script will automatically change the pref file value of ExistingV to yes from no once the process has finished.
Once the process has completed, additional files with the label unstripped should be generated inside the calibration folder, if ExistingV is no.
Example - General Script Utilised To Process Powder Diffraction With Polaris
from mantid.simpleapi import *
from mantid import config
import cry_ini
import cry_focus
# Browse to the directory of the instrument and move the instrument directory up to the top
# with the use of Move Up button
DIRS = config['datasearch.directories'].split(';')
# Alternatively you could also pass the path where the instrument folder is located
# DIRS = X:\
expt = cry_ini.Files('Polaris', RawDir=(DIRS[0] + "Polaris"), Analysisdir='test', forceRootDirFromScripts=False, inputInstDir=DIRS[0])
expt.initialize('Cycle_15_2', user='Mantid_tester', prefFile='UserPrefFile_15_2.pref')
expt.tell()
cry_focus.focus_all(expt, "79514", Write_ExtV=False)