Table of Contents
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| InputWorkspace1 | InOut | MatrixWorkspace | Mandatory | The name of the first input workspace |
| InputWorkspace2 | Input | MatrixWorkspace | Mandatory | The name of the second input workspace |
| CheckOverlapping | Input | boolean | True | Verify that the supplied data do not overlap |
| YAxisLabel | Input | string | The label to set the Y axis to | |
| YAxisUnit | Input | string | The unit to set the Y axis to |
This algorithm can be useful when working with large datasets. It enables the raw file to be loaded in two parts (not necessarily of equal size), the data processed in turn and the results joined back together into a single dataset. This can help avoid memory problems either because intermediate workspaces will be smaller and/or because the data will be much reduced after processing.
The output of the algorithm, in which the data from the second input workspace will be appended to the first, will be stored under the name of the first input workspace. Workspace data members other than the data (e.g. instrument etc.) will be copied from the first input workspace (but if they’re not identical anyway, then you probably shouldn’t be using this algorithm!). Both input workspaces will be deleted.
| Example case with input workspaces having 2 and 3 spectra respectively. | 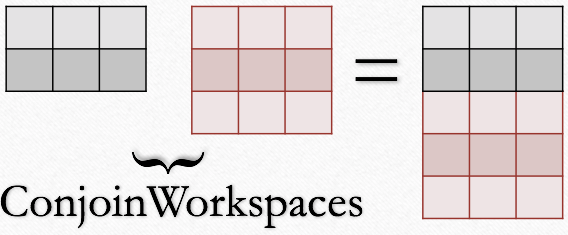
|
The algorithm adds the spectra from the first workspace and then the second workspace.
The input workspaces must come from the same instrument, have common units and bins and no detectors that contribute to spectra should overlap.
The optional parameters YAxisUnit and YAxisLabel can be used to change the y axis unit and label when conjoining workspaces. Changing YAxisUnit updates YAxisLabel automatically with the value of YAxisUnit, unless a separate value is supplied.
ConjoinWorkspaces Example
ws1 = CreateSampleWorkspace(WorkspaceType="Histogram", NumBanks=2, BankPixelWidth=1, BinWidth=10, Xmax=50)
print("Number of spectra in first workspace = {}".format(ws1.getNumberHistograms()))
ws2 = CreateSampleWorkspace(WorkspaceType="Histogram", NumBanks=3, BankPixelWidth=1, BinWidth=10, Xmax=50)
print("Number of spectra in second workspace = {}".format(ws2.getNumberHistograms()))
ConjoinWorkspaces(InputWorkspace1=ws1, InputWorkspace2=ws2, CheckOverlapping=False, YAxisUnit="New unit", YAxisLabel="New label")
ws = mtd['ws1'] # Have to update workspace from ADS, as it is an in-out parameter
print("Number of spectra after ConjoinWorkspaces = {}".format(ws.getNumberHistograms()))
print("Y unit is {}".format(ws.YUnit()))
print("Y label {}".format(ws.YUnitLabel()))
Output:
Number of spectra in first workspace = 2
Number of spectra in second workspace = 3
Number of spectra after ConjoinWorkspaces = 5
Y unit is New unit
Y label New label
Categories: Algorithm Index | Transforms\Merging
C++ source: ConjoinWorkspaces.cpp (last modified: 2018-12-16)
C++ header: ConjoinWorkspaces.h (last modified: 2018-10-05)