Table of Contents
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| Filenames | Input | list of str lists | Mandatory | Select one or more DNS SCD .d_dat files to load.Files must be measured at the same conditions. Allowed extensions: [‘.d_dat’] |
| OutputWorkspace | Output | MDEventWorkspace | Mandatory | An output MDEventWorkspace. |
| NormalizationWorkspace | Output | MDEventWorkspace | Mandatory | An output normalization MDEventWorkspace. |
| Normalization | Input | string | monitor | Algorithm will create a separate normalization workspace. Choose whether it should contain monitor counts or time. Allowed values: [‘monitor’, ‘time’] |
| a | Input | number | 1 | Lattice parameter a in Angstrom |
| b | Input | number | 1 | Lattice parameter b in Angstrom |
| c | Input | number | 1 | Lattice parameter c in Angstrom |
| alpha | Input | number | 90 | Angle between b and c in degrees |
| beta | Input | number | 90 | Angle between a and c in degrees |
| gamma | Input | number | 90 | Angle between a and b in degrees |
| OmegaOffset | Input | number | 0 | Angle in degrees between (HKL1) and the beam axisif the goniometer is at zero. |
| HKL1 | Input | dbl list | 1,1,0 | Indices of the vector in reciprocal space in the horizontal plane at angle Omegaoffset, if the goniometer is at zero. |
| HKL2 | Input | dbl list | 0,0,1 | Indices of a second vector in reciprocal space in the horizontal plane not parallel to HKL1 |
| TwoThetaLimits | Input | dbl list | 0,180 | Range (min, max) of scattering angles (2theta, in degrees) to consider. Everything out of this range will be cut. |
| LoadHuberFrom | Input | TableWorkspace | A table workspace to load a list of raw sample rotation angles. Huber angles given in the data files will be ignored. | |
| SaveHuberTo | Output | TableWorkspace | A workspace name to save a list of raw sample rotation angles. | |
| ElasticChannel | Input | number | 0 | Elastic channel number. Only for TOF data. |
| DeltaEmin | Input | number | -10 | Minimal energy transfer to consider. Should be <=0. Only for TOF data. |
Warning
This algorithm does not perform any consistency check of the input data. It is the users responsibility to choose a physically reasonable dataset.
This algorithm loads a list of DNS .d_dat data files into a MDEventWorkspace. If the algorithm fails to process a file, this file will be ignored. In this case the algorithm produces a warning and continues to process further files. Only if no valid files are provided, the algorithm terminates with an error message.
This algorithm is meant to replace the LoadDNSLegacy v1 for single crystal diffraction data.
Output
As a result, two workspaces are created:
Both workspaces have 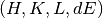 dimensions. The metadata are loaded into time series sample logs.
dimensions. The metadata are loaded into time series sample logs.
Note
For the further data reduction BinMD v1 should be used to bin the data.
For standard data (vanadium, NiCr, background) the sample rotation angle is assumed to be not important. These data are typically measured only for one sample rotation angle. The algorithm can replicate these data for the same sample rotation angles as a single crystal sample has been measured. For this purpose optional input fields SaveHuberTo and LoadHuberFrom can be used.
Note
If LoadHuberFrom option is applied, sample rotation angles in the data files will be ignored. Only sample rotation angles from the table will be considered.
Example 1 - Load a DNS .d_dat file to MDEventWorkspace:
# data file.
filename = "dn134011vana.d_dat"
# lattice parameters
a = 5.0855
b = 5.0855
c = 14.0191
omega_offset = 225.0
hkl1="1,0,0"
hkl2="0,0,1"
alpha=90.0
beta=90.0
gamma=120.0
# load data to MDEventWorkspace
ws, ws_norm, huber_ws = LoadDNSSCD(FileNames=filename, NormalizationWorkspace='ws_norm',
Normalization='monitor', a=a, b=b, c=c, alpha=alpha, beta=beta, gamma=gamma,
OmegaOffset=omega_offset, HKL1=hkl1, HKL2=hkl2, SaveHuberTo='huber_ws')
# print output workspace information
print("Output Workspace Type is: {}".format(ws.id()))
print("It has {0} events and {1} dimensions:".format(ws.getNEvents(), ws.getNumDims()))
for i in range(ws.getNumDims()):
dimension = ws.getDimension(i)
print("Dimension {0} has name: {1}, id: {2}, Range: {3:.2f} to {4:.2f} {5}".format(i,
dimension.getDimensionId(),
dimension.name,
dimension.getMinimum(),
dimension.getMaximum(),
dimension.getUnits()))
# print information about the table workspace
print ("TableWorkspace '{0}' has {1} row in the column '{2}'.".format(huber_ws.name(),
huber_ws.rowCount(),
huber_ws.getColumnNames()[0]))
print("It contains sample rotation angle {} degrees".format(huber_ws.cell(0, 0)))
Output:
Output Workspace Type is: MDEventWorkspace<MDEvent,4>
It has 24 events and 4 dimensions:
Dimension 0 has name: H, id: H, Range: -15.22 to 15.22 r.l.u
Dimension 1 has name: K, id: K, Range: -15.22 to 15.22 r.l.u
Dimension 2 has name: L, id: L, Range: -41.95 to 41.95 r.l.u
Dimension 3 has name: DeltaE, id: DeltaE, Range: -10.00 to 4.64 r.l.u
TableWorkspace 'huber_ws' has 1 row in the column 'Huber(degrees)'.
It contains sample rotation angle 79.0 degrees
Example 2 - Specify scattering angle limits:
# data file.
filename = "dn134011vana.d_dat"
# lattice parameters
a = 5.0855
b = 5.0855
c = 14.0191
omega_offset = 225.0
hkl1="1,0,0"
hkl2="0,0,1"
alpha=90.0
beta=90.0
gamma=120.0
# scattering angle limits, degrees
tth_limits = "20,70"
# load data to MDEventWorkspace
ws, ws_norm, huber_ws = LoadDNSSCD(FileNames=filename, NormalizationWorkspace='ws_norm',
Normalization='monitor', a=a, b=b, c=c, alpha=alpha, beta=beta, gamma=gamma,
OmegaOffset=omega_offset, HKL1=hkl1, HKL2=hkl2, TwoThetaLimits=tth_limits)
# print output workspace information
print("Output Workspace Type is: {}".format(ws.id()))
print("It has {0} events and {1} dimensions.".format(ws.getNEvents(), ws.getNumDims()))
# print normalization workspace information
print("Normalization Workspace Type is: {}".format(ws_norm.id()))
print("It has {0} events and {1} dimensions.".format(ws_norm.getNEvents(), ws_norm.getNumDims()))
Output:
Output Workspace Type is: MDEventWorkspace<MDEvent,4>
It has 10 events and 4 dimensions.
Normalization Workspace Type is: MDEventWorkspace<MDEvent,4>
It has 10 events and 4 dimensions.
Example 3 - Load sample rotation angles from the table
# data file.
filename = "dn134011vana.d_dat"
# construct table workspace with 10 raw sample rotation angles from 70 to 170 degrees
table = CreateEmptyTableWorkspace()
table.addColumn( "double", "Huber(degrees)")
for huber in range(70, 170, 10):
table.addRow([huber])
# lattice parameters
a = 5.0855
b = 5.0855
c = 14.0191
omega_offset = 225.0
hkl1="1,0,0"
hkl2="0,0,1"
alpha=90.0
beta=90.0
gamma=120.0
# load data to MDEventWorkspace
ws, ws_norm, huber_ws = LoadDNSSCD(FileNames=filename, NormalizationWorkspace='ws_norm',
Normalization='monitor', a=a, b=b, c=c, alpha=alpha, beta=beta, gamma=gamma,
OmegaOffset=omega_offset, HKL1=hkl1, HKL2=hkl2, LoadHuberFrom=table)
# print output workspace information
print("Output Workspace Type is: {}".format(ws.id()))
print("It has {0} events and {1} dimensions.".format(ws.getNEvents(), ws.getNumDims()))
# setting for the BinMD algorithm
bvec0 = '[100],unit,1,0,0,0'
bvec1 = '[001],unit,0,0,1,0'
bvec2 = '[010],unit,0,1,0,0'
bvec3 = 'dE,meV,0,0,0,1'
extents = '-2,1.5,-0.2,6.1,-10,10,-10,4.6'
bins = '10,10,1,1'
# bin the data
data_raw = BinMD(ws, AxisAligned='0', BasisVector0=bvec0, BasisVector1=bvec1, BasisVector2=bvec2,
BasisVector3=bvec3, OutputExtents=extents, OutputBins=bins, NormalizeBasisVectors='0')
# bin normalization
data_norm = BinMD(ws_norm, AxisAligned='0', BasisVector0=bvec0, BasisVector1=bvec1, BasisVector2=bvec2,
BasisVector3=bvec3, OutputExtents=extents, OutputBins=bins, NormalizeBasisVectors='0')
# normalize data
data = data_raw/data_norm
# print reduced workspace information
print("Reduced Workspace Type is: {}".format(data.id()))
print("It has {} dimensions.".format(data.getNumDims()))
s = data.getSignalArray()
print("Signal at some points: {0:.4f}, {1:.4f}, {2:.4f}".format(
float(s[7,1][0]), float(s[7,2][0]), float(s[7,3][0])))
Output:
Output Workspace Type is: MDEventWorkspace<MDEvent,4>
It has 240 events and 4 dimensions.
Reduced Workspace Type is: MDHistoWorkspace
It has 4 dimensions.
Signal at some points: 0.0035, 0.0033, 0.0035
Categories: Algorithm Index | MDAlgorithms\DataHandling
C++ source: LoadDNSSCD.cpp (last modified: 2018-12-14)
C++ header: LoadDNSSCD.h (last modified: 2018-10-05)