Table of Contents
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| SampleWorkspace | Input | MatrixWorkspace | Mandatory | Name of the Sample input Workspace |
| ResolutionWorkspace | Input | MatrixWorkspace | Mandatory | Name of the resolution input Workspace |
| EMin | Input | number | -0.2 | The start of the fitting range |
| EMax | Input | number | 0.2 | The end of the fitting range |
| SampleBins | Input | number | 1 | The number of sample bins |
| Elastic | Input | boolean | True | Fit option for using the elastic peak |
| Background | Input | string | Flat | Fit option for the type of background. Allowed values: [‘Sloping’, ‘Flat’, ‘Zero’] |
| NumberSigma | Input | number | 50 | Number of sigma values |
| NumberBeta | Input | number | 30 | Number of beta values |
| Loop | Input | boolean | True | Switch Sequential fit On/Off |
| OutputWorkspaceFit | Output | WorkspaceGroup | Mandatory | The name of the fit output workspaces |
| OutputWorkspaceContour | Output | WorkspaceGroup | Mandatory | The name of the contour output workspaces |
This is a variation of the stretched exponential option of
Quasi. For each spectrum a fit is performed
for a grid of 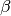 and
and 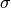 values. The distribution of goodness of fit values
is plotted.
values. The distribution of goodness of fit values
is plotted.
This routine was originally part of the MODES package. Note that this algorithm uses F2Py and is currently only supported on Windows.
Example - BayesStretch
# Check OS support for F2Py (Windows only)
from IndirectImport import is_supported_f2py_platform
if is_supported_f2py_platform():
# Load in test data
sample_ws = Load('irs26176_graphite002_red.nxs')
resolution_ws = Load('irs26173_graphite002_red.nxs')
# Run BayesStretch algorithm
fit_group, contour_group = BayesStretch(SampleWorkspace=sample_ws, ResolutionWorkspace=resolution_ws,
EMin=-0.2, EMax=0.2, Background='Sloping', Loop=True)
Categories: AlgorithmIndex | Workflow\MIDAS
Python: BayesStretch.py (last modified: 2019-02-26)