Table of Contents
import PyQt4
import mantid.simpleapi as simpleapi
import mantidqtpython as mpy
import sys
SpectrumView = mpy.MantidQt.SpectrumView.SpectrumView
app = PyQt4.QtGui.QApplication(sys.argv)
wsOut = simpleapi.Load("/Path/to/multispectrum/dataset")
sv = SpectrumView()
sv.renderWorkspace("wsOut")
sv.show()
app.exec_()
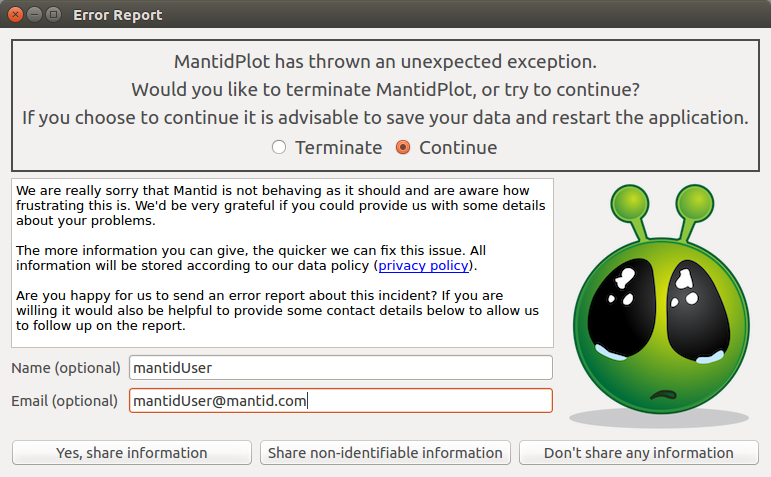
Error reporting has been enabled in place of the new last chance error handler. If Mantid catches an unknown exception it will now display the dialog box below. Currently there is no automatic error reporting enabled if Mantid crashes to desktop but the same system is planned to be implemented in this case as well.
The three options do the following:
Don’t share any information
The dialog box will close having sent no information. Mantid will either continue or terminate depending on which option has been selected at the top of the dialog.
Share non-identifiable information
An error report will be sent to errorreports.mantidproject.org. It will contain the following information:
Yes, share information
All the information from the non-identifiable information will be shared. In addition the optional name and email will be shared if given.
Full details of the privacy policy are available from the homepage.