\(\renewcommand\AA{\unicode{x212B}}\)
Table of Contents
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| Filename | Input | list of str lists | Files to load. Allowed extensions: [‘.nxs.h5’] | |
| IPTS | Input | number | Optional | IPTS number to load from |
| RunNumbers | Input | long list | Run numbers to load | |
| Grouping | Input | string | None | Group pixels. Allowed values: [‘None’, ‘2x2’, ‘4x4’] |
| OutputWorkspace | Output | Workspace | Mandatory | Output Workspace |
This algorithm will load a series of run into a MDHistoWorkspace that has dimensions x and y detector pixels vs scanIndex. The scanIndex is the omega rotation of the sample. The instrument from the first run only will be copied to the OutputWorkspace. In addition the s1 (omega rotation), duration, run_number and monitor count is read from every file and included in the logs of the OutputWorkspace.
If the “HB2C:CS:CrystalAlign:UBMatrix” property exist it will be converted into the OrientedLattice on the OutputWorkspace. The goniometer tilts (sgu and sgl) are combined into the UB Matrix so that only omega (s1) needs to be taken into account during rotation.
This algorithm doesn’t use Mantid loaders but instead h5py and numpy to load and integrate the events.
There is a grouping option to group pixels by either 2x2 or 4x4 which will help in reducing memory usage and speed up the later reduction steps. In most cases you will not see a difference in reduced data with 4x4 pixel grouping.
The loaded workspace is designed to be the input to ConvertWANDSCDtoQ v1.
Load one file, Vanadium for normalisation
norm = LoadWANDSCD(IPTS=7776, RunNumbers=26509)
print(repr(norm))
Output:
MDHistoWorkspace
Title:
Dim 0: (y) 0.5 to 512.5 in 512 bins
Dim 1: (x) 0.5 to 3840.5 in 3840 bins
Dim 2: (scanIndex) 0.5 to 1.5 in 1 bins
Inelastic: ki-kf
Instrument: ...
Run start: 2018-Mar-12 17:10:59
Run end: not available
Sample: a 1.0, b 1.0, c 1.0; alpha 90, beta 90, gamma 90
Load multiple data file
data = LoadWANDSCD(IPTS=7776, RunNumbers='26640-27944')
print("Memory used: {}GiB".format(data.getMemorySize()/2**30))
print(repr(data))
print('s1 = {}'.format(data.getExperimentInfo(0).run().getProperty('s1').value[0:10]))
print('monitor_counts = {}'.format(data.getExperimentInfo(0).run().getProperty('monitor_counts').value[0:10]))
print('duration = {}'.format(data.getExperimentInfo(0).run().getProperty('duration').value[0:10]))
print('run_number = {}'.format(data.getExperimentInfo(0).run().getProperty('run_number').value[0:10]))
Output:
Memory used: 59GB
MDHistoWorkspace
Title:
Dim 0: (y) 0.5 to 512.5 in 512 bins
Dim 1: (x) 0.5 to 3840.5 in 3840 bins
Dim 2: (scanIndex) 0.5 to 1305.5 in 1305 bins
Inelastic: ki-kf
Instrument: ...
Run start: 2018-May-02 13:34:10
Run end: not available
Sample: a 5.7, b 5.7, c 5.6; alpha 93, beta 90, gamma 98
s2 = [-180,-179.9,-179.8,-179.7,-179.6,-179.5,-179.4,-179.3,-179.2,-179.1]
monitor_count = [44571,44598,44567,44869,44453,44238,44611,44120,44762,44658]
duration = [2.05,2.05,2.03333,2.05,2.03333,2.03333,2.05,2.01667,2.05,2.05]
run_number = [26640,26641,26642,26643,26644,26645,26646,26647,26648,26649]
Load with different grouping comparing memory usage
data = LoadWANDSCD(IPTS=7776, RunNumbers='26640-27944')
data_2x2 = LoadWANDSCD(IPTS=7776, RunNumbers='26640-27944', Grouping='2x2')
data_4x4 = LoadWANDSCD(IPTS=7776, RunNumbers='26640-27944', Grouping='4x4')
print("Memory used by {}: {}GiB".format(data,data.getMemorySize()/2**30))
print("Memory used by {}: {}GiB".format(data_2x2,data_2x2.getMemorySize()/2**30))
print("Memory used by {}: {}GiB".format(data_4x4,data_4x4.getMemorySize()/2**30))
print(repr(data_4x4))
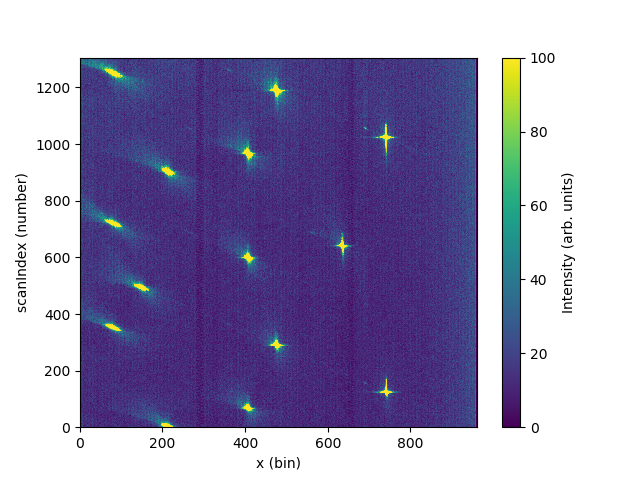
# Integrate y and plot
data_integrated = IntegrateMDHistoWorkspace('data_4x4', P1Bin='0,129')
import matplotlib.pyplot as plt
from mantid import plots
fig, ax = plt.subplots(subplot_kw={'projection':'mantid'})
c = ax.pcolormesh(data_integrated, vmax=100)
cbar=fig.colorbar(c)
cbar.set_label('Intensity (arb. units)')
#fig.savefig('LoadWANDSCD.png')
Output:
Memory used by data: 59GiB
Memory used by data_2x2: 14GiB
Memory used by data_4x4: 3GiB
MDHistoWorkspace
Title:
Dim 0: (y) 0.5 to 128.5 in 128 bins
Dim 1: (x) 0.5 to 960.5 in 960 bins
Dim 2: (scanIndex) 0.5 to 1305.5 in 1305 bins
Inelastic: ki-kf
Instrument: ...
Run start: 2018-May-02 13:34:10
Run end: not available
Sample: a 5.7, b 5.7, c 5.6; alpha 93, beta 90, gamma 98
Categories: AlgorithmIndex | DataHandling\Nexus
Python: LoadWANDSCD.py (last modified: 2020-03-27)