\(\renewcommand\AA{\unicode{x212B}}\)
Table of Contents
Save a table workspace containing fit parameters from EnggFitPeaks to an HDF5 file, indexed by bank ID
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| InputWorkspaces | Input | str list | Table workspaces containing fit parameters to save | |
| BankIDs | Input | long list | The bank ID of each input workspace, in order | |
| RunNumbers | Input | long list | The run number of each input workspace, in order | |
| Filename | Input | string | Mandatory | HDF5 file to save to. Allowed extensions: [‘.hdf5’, ‘.h5’, ‘.hdf’] |
Exports the results of an EnggFitPeaks fit to an HDF5 file indexed by bank ID. If multiple sets of fit results are provided, then the file is divided into sub-groups for each run number, with the Run Number groups indexed further by bank ID. The results go in a sub-group of the Bank group called Single Peak Fitting. This group contains one dataset, which is a table of the fit parameters for every peak
Obtaining fit results for banks 1 and 2 of run 123456 and then saving them with this algorithm would yield the following file structure:
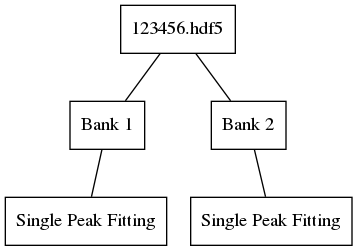
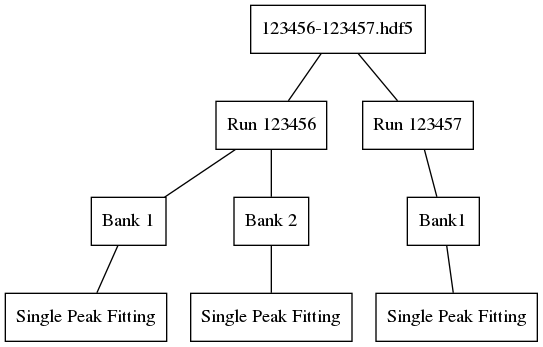
Obtaining fit results for banks 1 and 2 of run 123456 and bank 1 of 123457, and then saving them with this algorithm, would yield the following file structure:
The fit function used in EnggFitPeaks is Back2BackExponential, so the input table workspace must
contain values for the following parameters: ["dSpacing", "A0",
"A0_Err", "A1", "A1_Err", "X0", "X0_Err", "A", "A_Err", "B", "B_Err",
"S", "S_Err", "I", "I_Err", "Chi"]. Conveniently, this is exactly
what you get out of EnggFitPeaks.
Ordinarily, we’d get our peak parameters table from EnggFitPeaks, but we just mock one up here. See EnggFitPeaks documentation for how to generate this table.
Example - Export fit params to a new HDF5 file:
import h5py
import os
peaks = CreateEmptyTableWorkspace()
fit_param_headers = ["dSpacing", "A0", "A0_Err", "A1", "A1_Err", "X0", "X0_Err",
"A", "A_Err", "B", "B_Err", "S", "S_Err", "I", "I_Err", "Chi"]
[peaks.addColumn("double", header) for header in fit_param_headers]
for i in range(3):
peaks.addRow([(i + 1.0) / (j + 1) for j in range(len(fit_param_headers))])
output_filename = os.path.join(config["defaultsave.directory"],
"EnggSaveSinglePeakFitResultsToHDF5DocTest.hdf5")
EnggSaveSinglePeakFitResultsToHDF5(InputWorkspaces=[peaks],
Filename=output_filename,
BankIDs=[1])
with h5py.File(output_filename, "r") as f:
bank_group = f["Bank 1"]
peaks_dataset = bank_group["Single Peak Fitting"]
print("Peaks dataset has {} rows, for our 3 peaks".format(len(peaks_dataset)))
print("First peak is at D spacing {}".format(peaks_dataset[0]["dSpacing"]))
print("Third peak X0 = {}".format(peaks_dataset[2]["X0"]))
Output:
Peaks dataset has 3 rows, for our 3 peaks
First peak is at D spacing 1.0
Third peak X0 = 0.5
Categories: AlgorithmIndex | DataHandling
Python: EnggSaveSinglePeakFitResultsToHDF5.py (last modified: 2020-03-27)