\(\renewcommand\AA{\unicode{x212B}}\)
Table of Contents
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| Filename | Input | list of str lists | Mandatory | Files to combine in reduction. Allowed extensions: [‘_event.nxs’, ‘.nxs.h5’, ‘.nxs’] |
| Background | Input | string | Background run. Allowed extensions: [‘_event.nxs’, ‘.nxs.h5’, ‘.nxs’] | |
| BackgroundScale | Input | number | 1 | The background will be scaled by this number before being subtracted. |
| FilterByTofMin | Input | number | Optional | Optional: To exclude events that do not fall within a range of times-of-flight. This is the minimum accepted value in microseconds. Keep blank to load all events. |
| FilterByTofMax | Input | number | Optional | Optional: To exclude events that do not fall within a range of times-of-flight. This is the maximum accepted value in microseconds. Keep blank to load all events. |
| MomentumMin | Input | number | Mandatory | Minimum value in momentum. |
| MomentumMax | Input | number | Mandatory | Maximum value in momentum. |
| GroupingFile | Input | string | A file that consists of lists of spectra numbers to group. See GroupDetectors. Allowed extensions: [‘.xml’, ‘.map’] | |
| NormaliseByCurrent | Input | boolean | True | Normalise the Solid Angle workspace by the proton charge. |
| LoadInstrument | Input | string | Load a different instrument IDF onto the data from a file. See LoadInstrument. Allowed extensions: [‘.xml’] | |
| DetCal | Input | string | Load an ISAW DetCal calibration onto the data from a file. See LoadIsawDetCal. Allowed extensions: [‘.detcal’] | |
| MaskFile | Input | string | Masking file for masking. Supported file format is XML and ISIS ASCII. See LoadMask. Allowed extensions: [‘.xml’, ‘.msk’] | |
| SphericalAbsorptionCorrection | Input | boolean | False | Apply Spherical Absorption correction using AnvredCorrection |
| LinearScatteringCoef | Input | number | Optional | Linear scattering coefficient in 1/cm if not set with SetSampleMaterial |
| LinearAbsorptionCoef | Input | number | Optional | Linear absorption coefficient at 1.8 Angstroms in 1/cm if not set with SetSampleMaterial |
| Radius | Input | number | Optional | Radius of the sample in centimeters |
| SolidAngleOutputWorkspace | Output | Workspace | Mandatory | Output Workspace for Solid Angle |
| FluxOutputWorkspace | Output | Workspace | Mandatory | Output Workspace for Flux |
This algorithm preprocesses an incoherent scattering sample (usually
Vanadium) for use with MDNormSCD. The input
filename follows the syntax from MultipleFileProperty. The resulting workspaces can be
saved with SaveNexus.
Corelli example using multiple files with masking
# Create a simple mask file
MaskFilename=mantid.config.getString("defaultsave.directory")+"MDNormSCDPreprocessIncoherent_mask.xml"
LoadEmptyInstrument(InstrumentName='CORELLI', OutputWorkspace='CORELLI')
# Missing banks
MaskBTP('CORELLI', Bank='1-6,29,30,62,63-68,91')
# End of tubes
MaskBTP('CORELLI', Pixel='1-15,242-256')
SaveMask('CORELLI', MaskFilename)
SA, Flux = MDNormSCDPreprocessIncoherent(Filename='CORELLI_28119-28123',
MomentumMin=2.5,
MomentumMax=10,
MaskFile=MaskFilename)
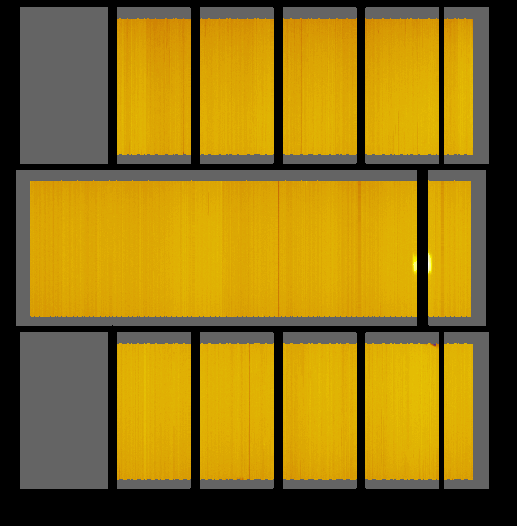
Solid Angle instrument view:
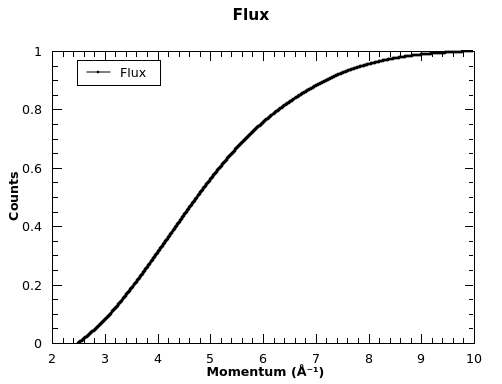
Flux spectrum plot:
TOPAZ example with backgound subtraction, grouping, masking and Spherical Absorption Correction
SA, Flux = MDNormSCDPreprocessIncoherent(Filename='TOPAZ_15670',
Background='TOPAZ_15671',
FilterByTofMin=500,
FilterByTofMax=16000,
MomentumMin=1.8,
MomentumMax=12.5,
GroupingFile='/SNS/TOPAZ/IPTS-15376/shared/calibration/TOPAZ_grouping_2016A.xml',
DetCal='/SNS/TOPAZ/IPTS-15376/shared/calibration/TOPAZ_2016A.DetCal',
MaskFile='/SNS/TOPAZ/IPTS-15376/shared/calibration/TOPAZ_masking_2016A.xml',
SphericalAbsorptionCorrection=True,
LinearScatteringCoef=0.367,
LinearAbsorptionCoef=0.366,
Radius=0.2)
print(SA.getNumberHistograms())
print(SA.blocksize())
print(Flux.getNumberHistograms())
print(Flux.blocksize())
Output:
1507328
1
23
10000
Python: MDNormSCDPreprocessIncoherent.py (last modified: 2020-03-27)