\(\renewcommand\AA{\unicode{x212B}}\)
Table of Contents
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| Filename | Input | list of str lists | Data files to load. Allowed extensions: [‘.dat’] | |
| IPTS | Input | number | Optional | IPTS number to load from |
| Exp | Input | number | Optional | Experiment number to load from |
| ScanNumbers | Input | long list | Scan numbers to load | |
| Vanadium | Input | string | Vanadium file, can be either the vanadium scan file or the reduced vcorr file. If not provided the vcorr file adjacent to the data file will be used. Allowed extensions: [‘.dat’, ‘.txt’] | |
| Normalise | Input | boolean | True | If False vanadium normalisation will not be performed |
| ExcludeDetectors | Input | long list | Detectors to exclude. If not provided the HB2A_exp???__exclude_detectors.txt adjacent to the data file will be used if it exist | |
| DefX | Input | string | By default the def_x (x-axis) from the file will be used, it can be overridden by setting it here | |
| IndividualDetectors | Input | boolean | False | If True the workspace will include each anode as a separate spectrum, useful for debugging issues |
| BinData | Input | boolean | True | Data will be binned using BinWidth. If False then all data will be unbinned |
| BinWidth | Input | number | 0.05 | Bin size of the output workspace |
| Scale | Input | number | 1 | The output will be scaled by this value |
| OutputWorkspace | Output | Workspace | Mandatory | Output Workspace |
| SaveData | Input | boolean | True | By default saving the reduced data to either GSAS or XYE |
| OutputFormat | Input | string | GSAS | Supportted output format: XYE (.dat), GSAS (.gss). Allowed values: [‘XYE’, ‘GSAS’] |
| OutputDirectory | Input | string | Saving directory for output file |
This algorithm reduces HFIR POWDER (HB-2A) data.
You can either specify the filenames of data you want to reduce or provide the IPTS, exp and scan number. If only one experiment exists in an IPTS then exp can be omitted. e.g. the following are equivalent:
ws = HB2AReduce('/HFIR/HB2A/IPTS-21073/exp666/Datafiles/HB2A_exp0666_scan0024.dat')
# and
ws = HB2AReduce(IPTS=21073, exp=666, ScanNumbers=24)
You can specify any number of filenames or scan numbers (in a comma separated list).
By default the correct vcorr file (HB2A_exp???__Ge_[113|115]_[IN|OUT]_vcorr.txt) adjacent to the data file will be used. Alternatively either the vcorr file or a vanadium scan file can be provided to the Vanadium option. If a vanadium scan file is provided then the vanadium counts can be taken into account when calculating the uncertainty which can not be done with using the vcorr file.
If Normalise=False then no normalisation will be performed.
By default the file HB2A_exp???__exclude_detectors.txt adjacent to the data file will be used unless a list of detectors to exclude are provided by ExcludeDetectors
If this option is True then a separate spectra will be created in the output workspace for every anode. This allows you to compare adjacent anodes.
If BinData=True (default) then the data will be binned on a regular grid with a width of BinWidth. The output can be scaled by an arbitrary amount by setting Scale.
This algorithm will read the def_x value in the data file and use it as the x-axis. This value can be overridden by setting the DefX property, e.g. DefX='2theta'.
If you did a scan using a particular anode vs temperature then you should set IndividualDetectors=True and specify DefX if not correct in the data file. Then simply plot the spectrum you are scanning, look at the example below Anode8 vs temperature.
The output workspace can be saved to XYE, Maud and TOPAS format using SaveFocusedXYE. e.g.
# XYE with no header
SaveFocusedXYE(ws, Filename='data.xye', SplitFiles=False, IncludeHeader=False)
# TOPAS format
SaveFocusedXYE(ws, Filename='data.xye', SplitFiles=False, Format='TOPAS')
# Maud format
SaveFocusedXYE(ws, Filename='data.xye', SplitFiles=False, Format='MAUD')
You can also save the reduced data as GSAS or XYE format by adding additional arguments to the reduction call
ws = HB2AReduce(
'/HFIR/HB2A/IPTS-21073/exp666/Datafiles/HB2A_exp0666_scan0024.dat',
SaveData=True,
OutputFormat="GSAS",
OutputDirectory="/tmp",
)
Warning
- Do not specify OutputFormat or OutputDirectory if SaveData is set to False.
- If def_x = 2theta is not the in the header of any one of the input files, do not set OutputFormat to GSAS.
Individual Detectors
ws=HB2AReduce('HB2A_exp0666_scan0024.dat', IndividualDetectors=True)
# Plot anodes 40, 41 and 42
import matplotlib.pyplot as plt
from mantid import plots
fig, ax = plt.subplots(subplot_kw={'projection':'mantid'})
for num in [40,41,42]:
ax.plot(ws, specNum=num)
plt.legend()
#fig.savefig('HB2AReduce_1.png')
plt.show()
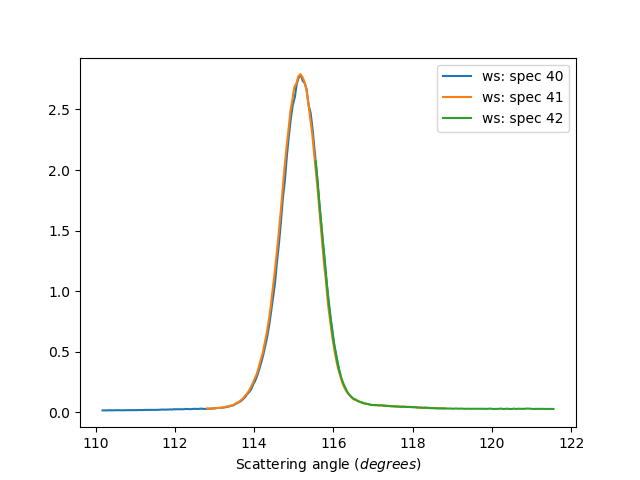
Unbinned data
ws=HB2AReduce('HB2A_exp0666_scan0024.dat', BinData=False)
# Plot
import matplotlib.pyplot as plt
from mantid import plots
fig, ax = plt.subplots(subplot_kw={'projection':'mantid'})
ax.plot(ws)
#fig.savefig('HB2AReduce_2.png')
plt.show()
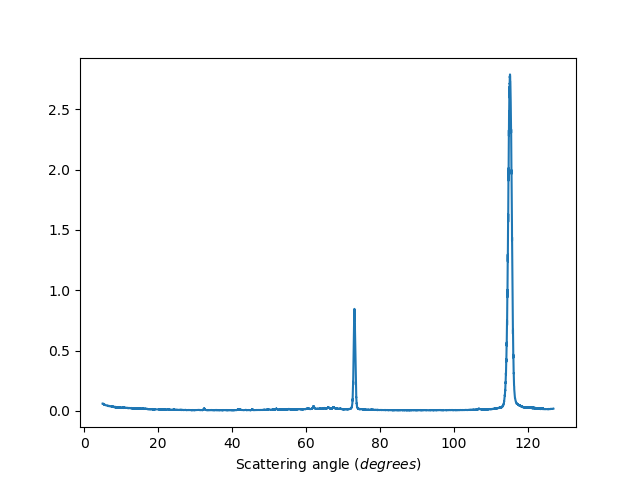
Binned data
ws=HB2AReduce('HB2A_exp0666_scan0024.dat')
# Plot
import matplotlib.pyplot as plt
from mantid import plots
fig, ax = plt.subplots(subplot_kw={'projection':'mantid'})
ax.plot(ws)
#fig.savefig('HB2AReduce_3.png')
plt.show()
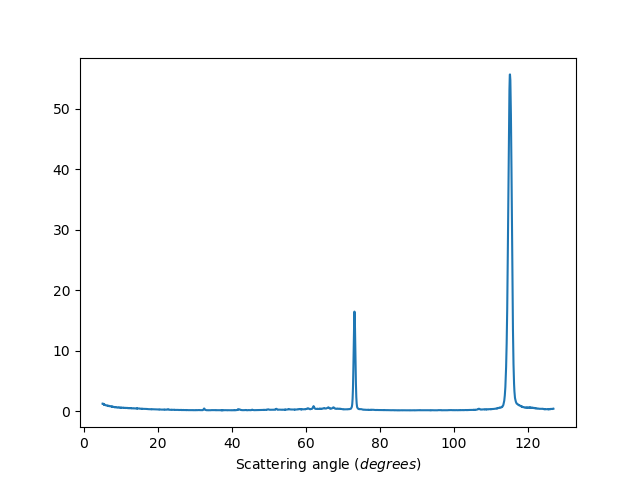
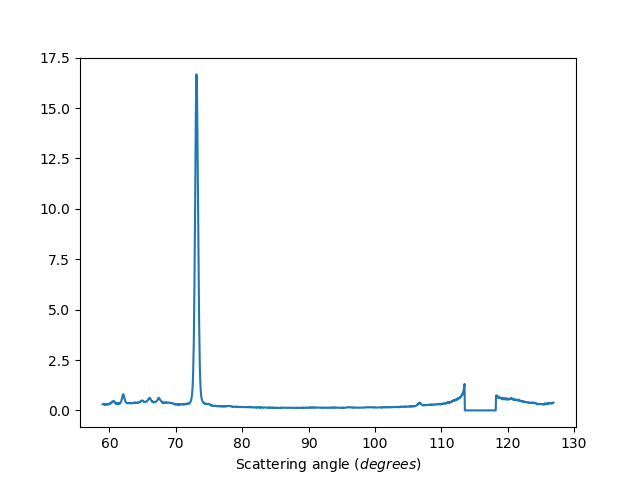
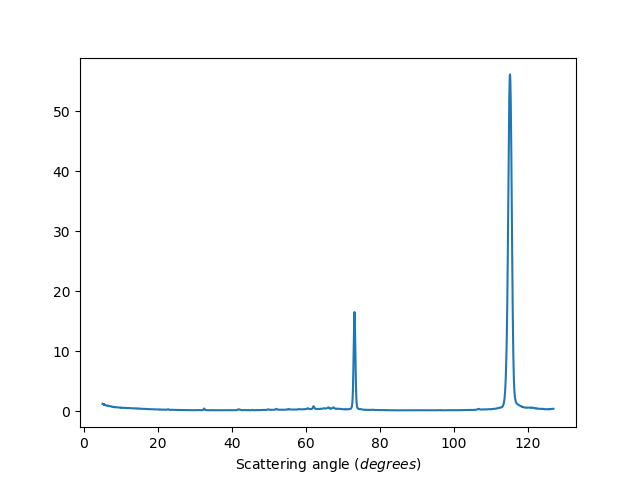
Exclude detectors: 1-20,40,41,42
ws=HB2AReduce('HB2A_exp0666_scan0024.dat', ExcludeDetectors='1-20,40,41,42')
# Plot
import matplotlib.pyplot as plt
from mantid import plots
fig, ax = plt.subplots(subplot_kw={'projection':'mantid'})
ax.plot(ws)
#fig.savefig('HB2AReduce_4.png')
plt.show()
Combining multiple files
ws=HB2AReduce('HB2A_exp0666_scan0024.dat, HB2A_exp0666_scan0025.dat')
# Plot
import matplotlib.pyplot as plt
from mantid import plots
fig, ax = plt.subplots(subplot_kw={'projection':'mantid'})
ax.plot(ws)
#fig.savefig('HB2AReduce_5.png')
plt.show()
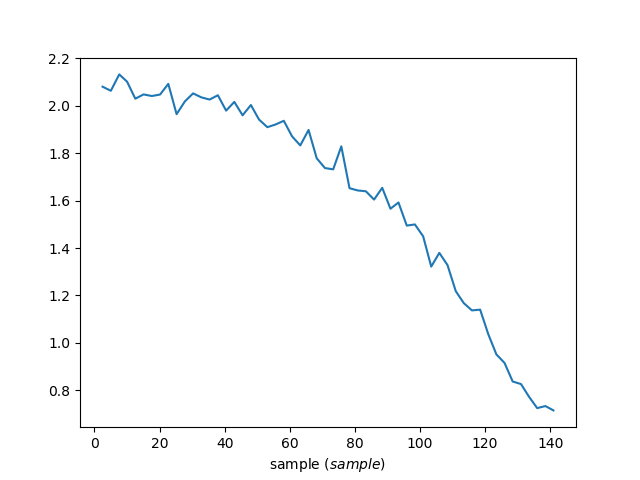
Anode8 vs temperature
Because the following data file has def_x = sample then this
algorithm will reduce the data to be counts vs sample (sample
temperature). Setting IndividualDetectors=True allows you to see a
single anode vs temperature.
ws=HB2AReduce('HB2A_exp0660_scan0146.dat',
Vanadium='HB2A_exp0644_scan0018.dat',
IndividualDetectors=True)
# Plot
import matplotlib.pyplot as plt
from mantid import plots
fig, ax = plt.subplots(subplot_kw={'projection':'mantid'})
ax.plot(ws, specNum=8) # anode8
#fig.savefig('HB2AReduce_6.png')
plt.show()
Categories: AlgorithmIndex | Diffraction\Reduction
Python: HB2AReduce.py