\(\renewcommand\AA{\unicode{x212B}}\)
CompactMD v1¶

Enable screenshots using DOCS_SCREENSHOTS in CMake¶
Summary¶
Crops an MDHistoWorkspace based on the first non-zero signals giving a more focussed area of interest.
Properties¶
Name |
Direction |
Type |
Default |
Description |
|---|---|---|---|---|
InputWorkspace |
Input |
IMDHistoWorkspace |
Mandatory |
MDHistoWorkspace to compact |
OutputWorkspace |
Output |
IMDHistoWorkspace |
Mandatory |
Output compacted workspace |
Description¶
Used to crop an n-dimensional MDHistoWorkspace to the first non-zero signal values found in all dimensions.
Cropping¶
The cropping is done by supplying IntegrateMDHistoWorkspace with the minimum and maximum extents associated with the first non-zero signal values in the workspace.
Usage¶
Example - CompactMD on MDHistoWorkspace
import math
#create an MDEventWorkspace for Rebinning
mdws = CreateMDWorkspace(Dimensions=3, Extents='-10,10,-10,10,-10,10', Names='A,B,C', Units='U,U,U')
FakeMDEventData(InputWorkspace=mdws, PeakParams='100000,-5,-5,0,1')
FakeMDEventData(InputWorkspace=mdws, PeakParams='100000,0,0,0,1')
FakeMDEventData(InputWorkspace=mdws, PeakParams='100000,5,5,0,1')
#Rebin mdws to create an MDHistoWorkspace
binned_ws = BinMD(InputWorkspace=mdws, AxisAligned=False, BasisVector0='a,unit,1,1,0',BasisVector1='b,unit,-1,1,0',BasisVector2='c,unit,0,0,1',NormalizeBasisVectors=True,Translation=[-10,-10,0], OutputExtents=[0,math.sqrt(2*20*20),-2,2,-10,10], OutputBins=[100, 100, 1] )
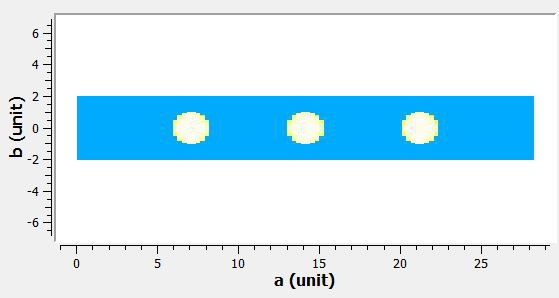
#A visualisation of the rebinned_ws can be found in the 'Input' section below.
#run CompactMD on the rebinned workspace
compact_output = CompactMD(binned_ws)
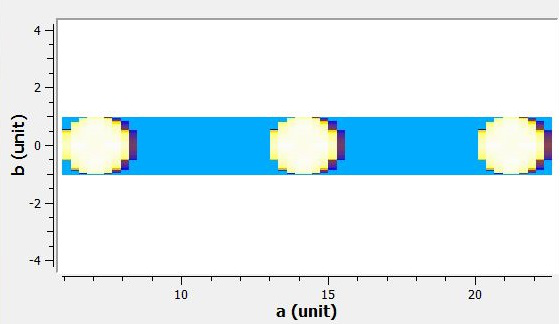
#A visualisation of the compacted workspace can be found in the 'Output' section below.
Input:
Output:
Categories: AlgorithmIndex | MDAlgorithms\Utility\Workspaces
Source¶
C++ header: CompactMD.h
C++ source: CompactMD.cpp
