\(\renewcommand\AA{\unicode{x212B}}\)
MDNormDirectSC v1¶

Enable screenshots using DOCS_SCREENSHOTS in CMake¶
Summary¶
Calculate normalization for an MDEvent workspace for single crystal direct geometry inelastic measurement.
See Also¶
Properties¶
Name |
Direction |
Type |
Default |
Description |
|---|---|---|---|---|
InputWorkspace |
Input |
Mandatory |
An input MDWorkspace. |
|
AlignedDim0 |
Input |
string |
Binning parameters for the 0th dimension. Enter it as a comma-separated list of values with the format: ‘name,minimum,maximum,number_of_bins’. Leave blank for NONE. |
|
AlignedDim1 |
Input |
string |
Binning parameters for the 1th dimension. Enter it as a comma-separated list of values with the format: ‘name,minimum,maximum,number_of_bins’. Leave blank for NONE. |
|
AlignedDim2 |
Input |
string |
Binning parameters for the 2th dimension. Enter it as a comma-separated list of values with the format: ‘name,minimum,maximum,number_of_bins’. Leave blank for NONE. |
|
AlignedDim3 |
Input |
string |
Binning parameters for the 3th dimension. Enter it as a comma-separated list of values with the format: ‘name,minimum,maximum,number_of_bins’. Leave blank for NONE. |
|
AlignedDim4 |
Input |
string |
Binning parameters for the 4th dimension. Enter it as a comma-separated list of values with the format: ‘name,minimum,maximum,number_of_bins’. Leave blank for NONE. |
|
AlignedDim5 |
Input |
string |
Binning parameters for the 5th dimension. Enter it as a comma-separated list of values with the format: ‘name,minimum,maximum,number_of_bins’. Leave blank for NONE. |
|
SolidAngleWorkspace |
Input |
An input workspace containing integrated vanadium (a measure of the solid angle). |
||
SkipSafetyCheck |
Input |
boolean |
False |
If set to true, the algorithm does not check history if the workspace was modified since theConvertToMD algorithm was run, and assume that the direct geometry inelastic mode is used. |
TemporaryNormalizationWorkspace |
Input |
IMDHistoWorkspace |
An input MDHistoWorkspace used to accumulate normalization from multiple MDEventWorkspaces. If unspecified a blank MDHistoWorkspace will be created. |
|
TemporaryDataWorkspace |
Input |
IMDHistoWorkspace |
An input MDHistoWorkspace used to accumulate data from multiple MDEventWorkspaces. If unspecified a blank MDHistoWorkspace will be created. |
|
OutputWorkspace |
Output |
Mandatory |
A name for the output data MDHistoWorkspace. |
|
OutputNormalizationWorkspace |
Output |
Mandatory |
A name for the output normalization MDHistoWorkspace. |
Description¶
The algorithm calculates a normalization MD workspace for single crystal direct geometry inelastic experiments. Trajectories of each detector in reciprocal space are calculated, and the flux is integrated between intersections with each MDBox. A brief introduction to the multi-dimensional data normalization can be found here.
Note
If the MDEvent input workspace is generated from an event workspace, the algorithm gives the correct normalization only if the event workspace is cropped and binned to the same energy transfer range. If the workspace is not cropped, one might have events in places where the normalization is calculated to be 0.
Note
As of Release 4.0.0, the algorithm can handle merged MD workspaces. Make sure all original MDEvent workspaces have the same dimensions
Usage¶
Note
To run these usage examples please first download the usage data, and add these to your path. In Mantid this is done using Manage User Directories.
Example - MDNormDirectSC
import mantid
import os
from mantid.simpleapi import *
config['default.facility']="SNS"
from numpy import *
DGS_input_data=Load("CNCS_7860")
# Keep events (SofPhiEIsDistribution=False)
# Do not normalize by proton charge in DgsReduction
DGS_output_data=DgsReduction(
SampleInputWorkspace=DGS_input_data,
SofPhiEIsDistribution=False,
IncidentBeamNormalisation="None",
EnergyTransferRange="-1.5,0.01,2.7",
)
DGS_output_data=DGS_output_data[0]
SetGoniometer(DGS_output_data,Axis0="10.,0,1,0,1")
SetUB(DGS_output_data, 5.,3.2,7.2)
DGS_output_data=CropWorkspace(DGS_output_data,XMin=-1.5,XMax=2.7)
MDE=ConvertToMD(DGS_output_data,
QDimensions="Q3D",
dEAnalysisMode="Direct",
Q3DFrames="HKL",
QConversionScales="HKL")
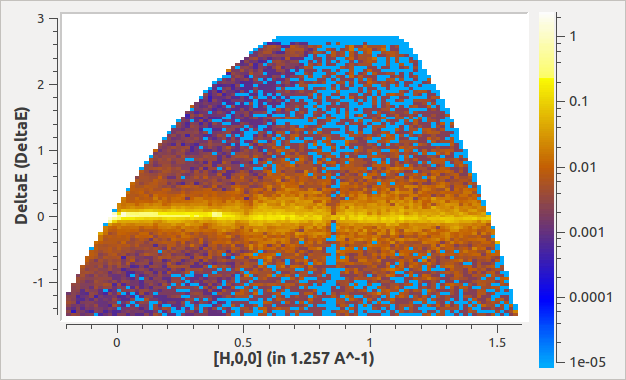
histoData,histoNorm=MDNormDirectSC(MDE,
AlignedDim0="[H,0,0],-0.2,1.6,100",
AlignedDim1="DeltaE,-1.5,3.,100",
)
normalized=histoData/histoNorm
histoShape=histoNorm.getSignalArray().shape
print "The normalization workspace shape is (%d, %d)" % histoShape
print "Out of those elements, "+str(nonzero(histoNorm.getSignalArray())[0].size)+" are nonzero"
The normalization workspace shape is (100, 100)
Out of those elements, 6712 are nonzero
The output would look like:
Categories: AlgorithmIndex | MDAlgorithms\Normalisation
Source¶
C++ header: MDNormDirectSC.h
C++ source: MDNormDirectSC.cpp
