SofTwoThetaTOF v1¶
Summary¶
Calculates the intensity as a function of scattering angle and time of flight.
Properties¶
Name |
Direction |
Type |
Default |
Description |
|---|---|---|---|---|
InputWorkspace |
Input |
Mandatory |
A workspace to be converted. |
|
OutputWorkspace |
Output |
Mandatory |
A workspace with (2theta, TOF) units. |
|
AngleStep |
Input |
number |
Optional |
The angle step for detector grouping, in degrees. |
GroupingFilename |
Input |
string |
A grouping file that will be created; a corresponding .par file wille be created as well. Allowed extensions: [‘.xml’] |
Description¶
This algorithm converts a workspace from (spectrum number, TOF) coordinates to (\(2\theta\), TOF). The latter view is useful for visualization and diagnostics purposes of direct geometry spectrometers with large PSD arrays. The algorithm performs the following steps:
Equalize the sample-to-detector distances using ConvertToConstantL2
Rebin the ragged workspace to same binning over all histograms. The bin width is taken from the first bin of the first histogram. This will introduce zero padding to the histograms.
Mask the zero padding using MaskNonOverlappingBins.
Group the detectors to given \(2\theta\) bins using GenerateGroupingPowder and GroupDetectors.
Convert the spectrum axis from spectrum numbers to \(2\theta\) using ConvertSpectrumAxis
A default AngleStep can be defined in the instrument parameters. The parameter is called natural-angle-step; its type should be float and value the desired AngleStep in degrees.
The instrument should have an l2 parameter defined which is the nominal sample to detector distance. Also, sample logs should include a wavelength entry (in Angstroms). See ConvertToConstantL2 for more details.
By default, the .xml and .par files generated by GenerateGroupingPowder are stored in a temporary location and deleted when the algorithm finishes. If these files are needed afterwards, a name for the .xml file can be given by GroupingFilename. The .par file will have the same name except for the file extension which is changed from .xml to .par. Note that this algorithm forces the .xml extension to GroupingFilename.
Usage¶
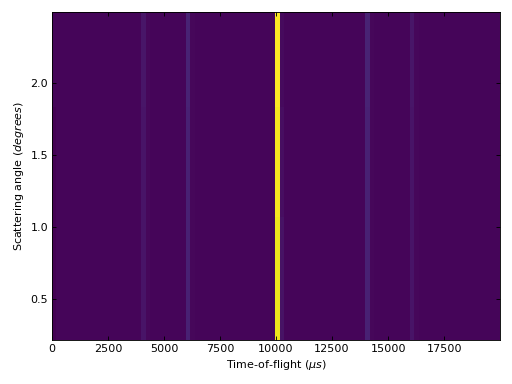
Example - Applying SofTwoThetaTOF and plotting the result
from mantid.simpleapi import *
import directtools as dt
ws = CreateSampleWorkspace(Function='Quasielastic Tunnelling', NumBanks=1, BankPixelWidth=20)
AddSampleLog(ws, 'wavelength', '6.26', 'Number', 'Angstrom')
SetInstrumentParameter(ws, ParameterName='l2', ParameterType='String', Value='5')
SofTT = SofTwoThetaTOF(ws, AngleStep=1)
fig, axes = dt.subplots()
axes.pcolor(SofTT)
axes.set_xlim(0.)
# Uncomment the line below to show the plot.
#fig.show()
(Source code, png, hires.png, pdf)
Categories: AlgorithmIndex | Inelastic | ILL\Direct
Source¶
C++ header: SofTwoThetaTOF.h
C++ source: SofTwoThetaTOF.cpp