Basic 1D, 2D and 3D Plots¶
plotSpectrum and plotBin¶
Right-clicking a workspace from the Workspaces Toolbox and selecting Show Data creates a Data Matrix, which displays the underlying data in a worksheet.
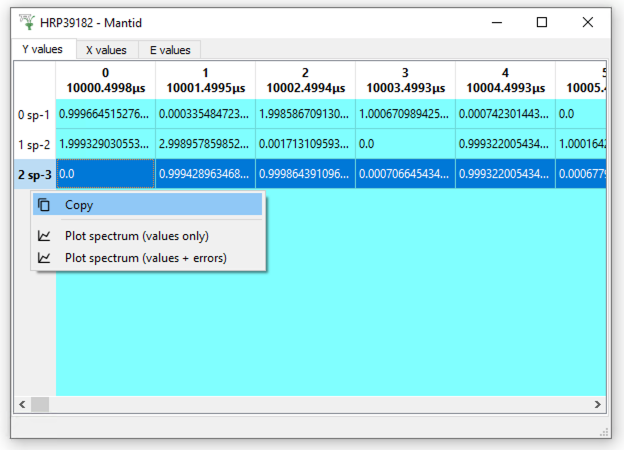
Right clicking on a row or a column allows a plot of that spectrum or time bin, which can also be achieved in Python using the plotSpectrum() or plotBin() functions, e.g.
RawData = Load("MAR11015")
graph_spec = plotSpectrum(RawData, 0)
graph_time = plotBin(RawData, 0)
To include error bars on a plot, there is an extra argument that can be set to ‘True’ to enable them, e.g.
graph_spec = plotSpectrum(RawData, 0, error_bars=True)
graph_time = plotBin(RawData, 0, error_bars=True)
For setting scales, axis titles, plot titles etc. you can use:
RawData = Load("MAR11015")
plotSpectrum(RawData,1, error_bars=True)
# Get current axes of the graph
fig, axes = plt.gcf(), plt.gca()
plt.yscale('log')
# Rescale the axis limits
axes.set_xlim(0,5000)
axes.set_ylim(0.001,1500)
#C hange the y-axis label
axes.set_ylabel(r'Counts ($\mu s$)$^{-1}$')
# Add legend entries
axes.legend(['Good Line'])
# Give the graph a modest title
plt.title("My Wonderful Plot", fontsize=20)
Multiple workspaces/spectra can be plotted by providing lists within the plotSpectrum() or plotBin() functions, e.g.
# Plot multiple spectra from a single workspace
plotSpectrum(RawData, [0,1,3])
# Here you need to load files into RawData1 and RawData2 before the next two commands
# Plot a spectrum from different workspaces
plotSpectrum([RawData1,RawData2], 0)
# Plot multiple spectra across multiple workspaces
plotSpectrum([RawData1,RawData2], [0,1,3])
To overplot on the same window:
RawData = Load("MAR11015")
# Assign original plot to a window called graph_spce
graph_spec = plotSpectrum(RawData, 0)
# Overplot on that window, without clearing it
plotSpectrum(RawData, 1, window=graph_spec, clearWindow=False)
2D Colourfill and Contour Plots¶
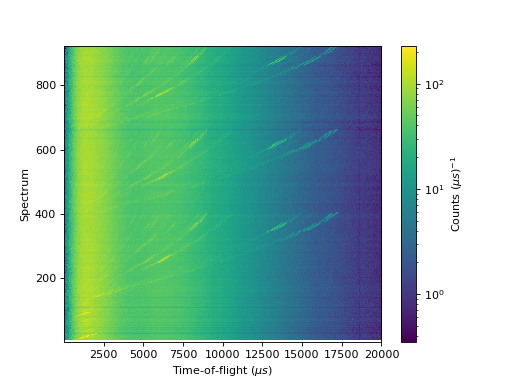
2D plots can be produced as an image or a pseudocolormesh (for a non-regular grid):
''' ----------- Image > imshow() ----------- '''
from mantid.simpleapi import *
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
data = Load('MAR11060')
fig, axes = plt.subplots(subplot_kw={'projection':'mantid'})
# IMPORTANT to set origin to lower
c = axes.imshow(data, origin = 'lower', cmap='viridis', aspect='auto', norm=LogNorm())
cbar=fig.colorbar(c)
cbar.set_label('Counts ($\mu s$)$^{-1}$') #add text to colorbar
#fig.show()
(Source code, png, hires.png, pdf)
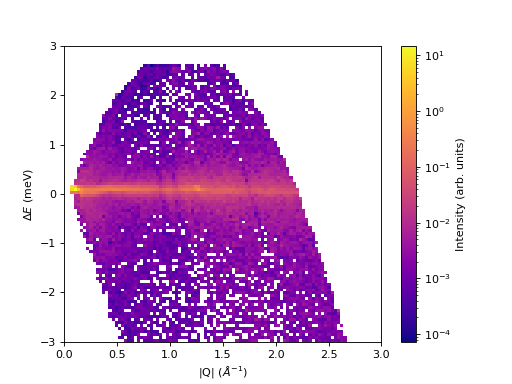
''' ----------- Pseudocolormesh > pcolormesh() ----------- '''
from mantid.simpleapi import *
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
data = Load('CNCS_7860')
data = ConvertUnits(InputWorkspace=data,Target='DeltaE', EMode='Direct', EFixed=3)
data = Rebin(InputWorkspace=data, Params='-3,0.025,3', PreserveEvents=False)
md = ConvertToMD(InputWorkspace=data,QDimensions='|Q|',dEAnalysisMode='Direct')
sqw = BinMD(InputWorkspace=md,AlignedDim0='|Q|,0,3,100',AlignedDim1='DeltaE,-3,3,100')
fig, ax = plt.subplots(subplot_kw={'projection':'mantid'})
c = ax.pcolormesh(sqw, cmap='plasma', norm=LogNorm())
cbar=fig.colorbar(c)
cbar.set_label('Intensity (arb. units)') #add text to colorbar
#fig.show()
(Source code, png, hires.png, pdf)
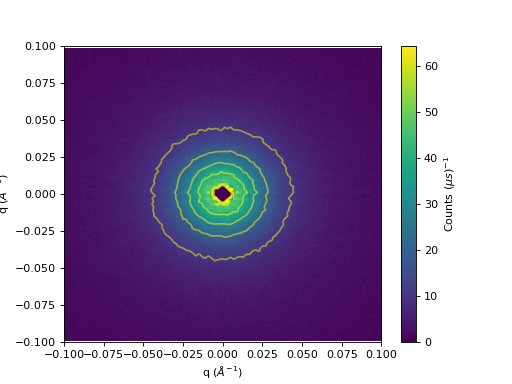
Contour lines can be overlayed on a 2D colorfill:
''' ----------- Contour overlay ----------- '''
from mantid.simpleapi import *
import matplotlib.pyplot as plt
import numpy as np
data = Load('SANSLOQCan2D.nxs')
fig, axes = plt.subplots(subplot_kw={'projection':'mantid'})
# IMPORTANT to set origin to lower
c = axes.imshow(data, origin = 'lower', cmap='viridis', aspect='auto')
# Overlay contours
axes.contour(data, levels=np.linspace(10, 60, 6), colors='yellow', alpha=0.5)
cbar=fig.colorbar(c)
cbar.set_label('Counts ($\mu s$)$^{-1}$') #add text to colorbar
#fig.show()
(Source code, png, hires.png, pdf)
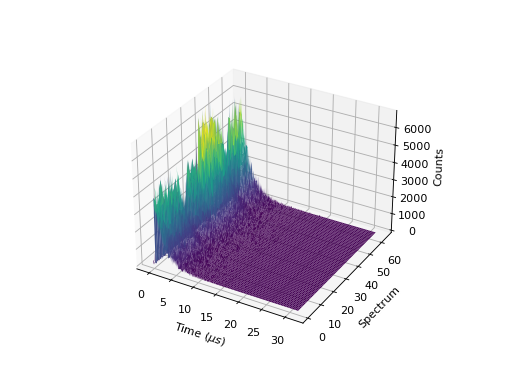
3D Surface and Wireframe Plots¶
3D plots Surface and Wireframe plots can also be created:
''' ----------- Surface plot ----------- '''
from mantid.simpleapi import *
import matplotlib.pyplot as plt
data = Load('MUSR00015189.nxs')
data = mtd['data_1'] # Extract individual workspace from group
fig, ax = plt.subplots(subplot_kw={'projection':'mantid3d'})
ax.plot_surface(data, cmap='viridis')
#fig.show()
(Source code, png, hires.png, pdf)
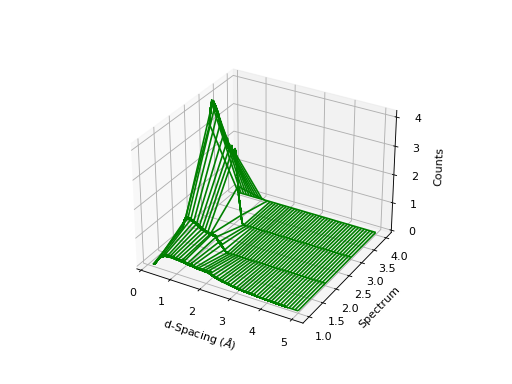
''' ----------- Wireframe plot ----------- '''
from mantid.simpleapi import *
import matplotlib.pyplot as plt
# This file can be found in the Usage Examples folder, available
# here https://www.mantidproject.org/installation/index#sample-data
data = Load('PG3_733.nxs')
fig, ax = plt.subplots(subplot_kw={'projection':'mantid3d'})
ax.plot_wireframe(data, color='green')
#fig.show()
(Source code, png, hires.png, pdf)
See here for custom color cycles and colormaps