MPI support in Mantid is based on processing a subset of spectra on each MPI rank. Many algorithms process each spectrum independently so this is an efficient way of splitting the computational effort and data volume.
Note that the multi-dimensional workspaces MDHistoWorkspace and MDEventWorkspace handle data differently and thus cannot be dealt with in this way. MPI support for these multi-dimensional workspaces is beyond the scope of this document.
In Mantid the terms spectrum and detector are used interchangeably at times, leading to unnecessary confusion. In particular, it is sometimes assumed that there is a 1:1 correspondence between spectra and detectors. It is important to clearly distinguish between a spectrum and a detector, especially in the context of MPI support. We may define the terms as follows:
Examples may help to clarify this:
For the purpose of MPI support, Mantid always stores the complete instrument including all detectors on every MPI rank. [1] Only a subset of all spectra is stored on the local MPI rank, so a detector may be locally without corresponding spectrum but have a spectrum associated to it on another MPI rank.
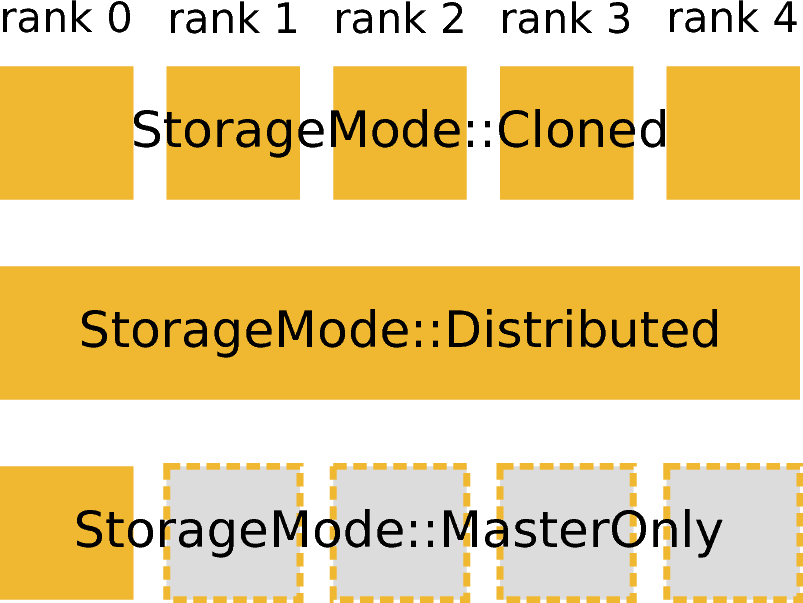
Visualization of available storage modes. In this and the following figures a vertical column is used to depict an MPI rank.
Not all data in an MPI-based data reduction can be or has to be distributed. Some data may be present on all ranks or only on a single rank. To formalize this concept a Workspace in Mantid now has an associated storage mode (Parallel::StorageMode), as visualized in the figure on the right. The storage mode of a workspace can be obtained by calling Workspace::storageMode(). There are three available storage modes
Currently only workspaces have an associated storage mode. For all other data, such as a scalar value provided as an input to an algorithm are implicitly assumed to have StorageMode::Cloned, i.e., have the same value on all MPI ranks.
Usage examples for the storage modes could include:
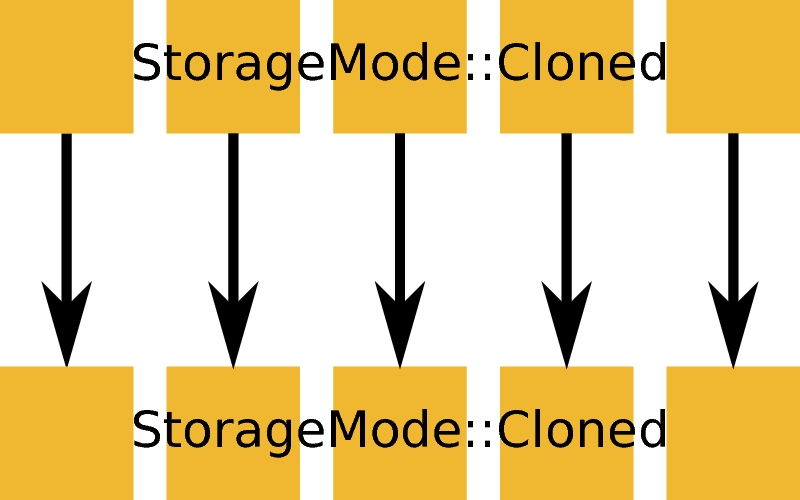
ExecutionMode::Identical based on an input and output workspace with StorageMode::Cloned. Example: ConvertUnits, Rebin, or many other algorithm that do not load or save data.
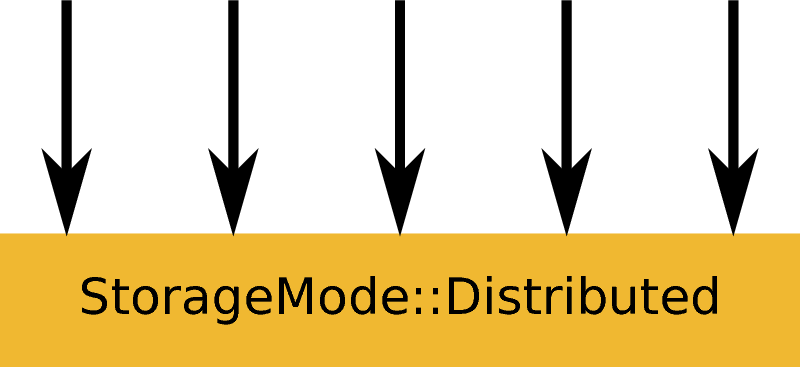
ExecutionMode::Distributed creating an output workspace with StorageMode::Distributed. Example: LoadEventNexus.
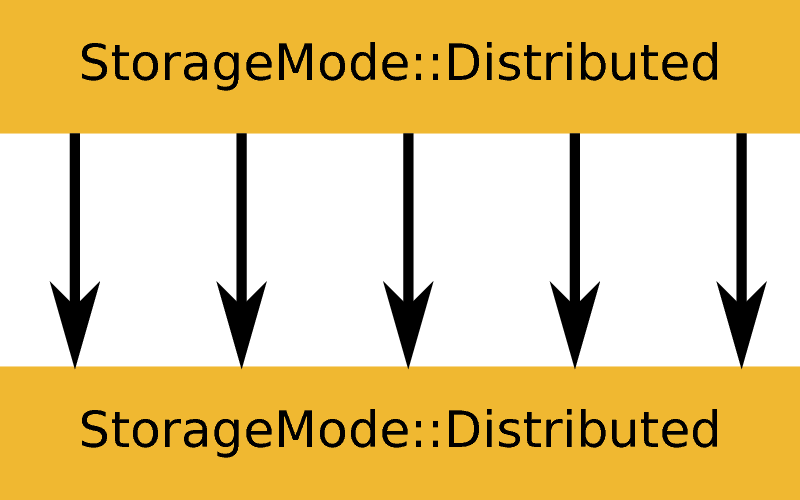
ExecutionMode::Distributed based on an input and output workspace with StorageMode::MasterOnly. Example: ConvertUnits or Rebin.
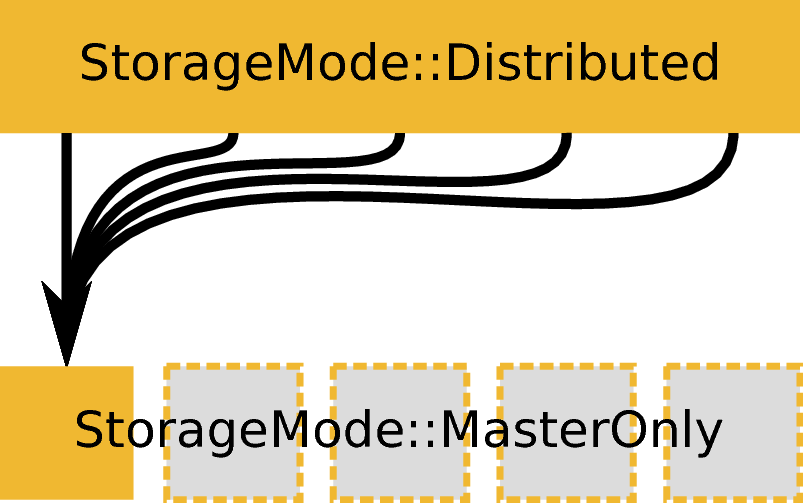
ExecutionMode::Distributed based on an input workspace with StorageMode::Distributed creating an output workspace with StorageMode::MasterOnly. Example: DiffractionFocussing.
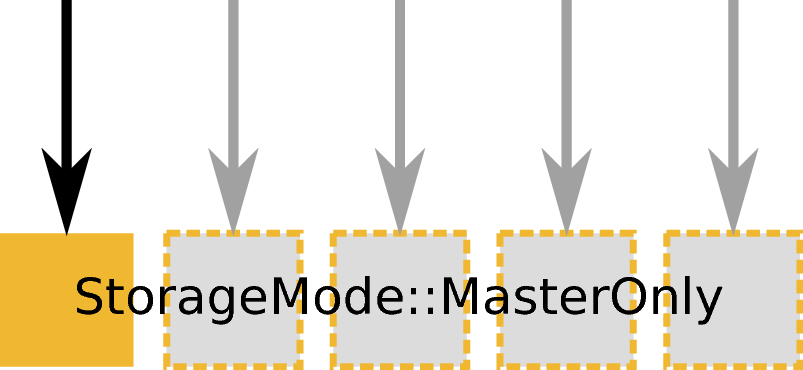
ExecutionMode::MasterOnly creating an output workspace with StorageMode::Distributed. Example: LoadEventNexus or other load algorithms.
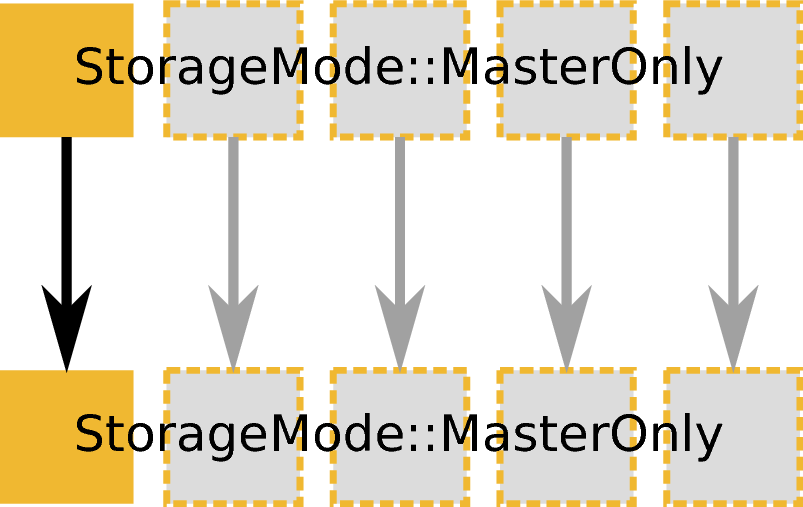
ExecutionMode::MasterOnly based on an input and output workspace with StorageMode::MasterOnly. Example: ConvertUnits, Rebin, or many other algorithm that do not load or save data.
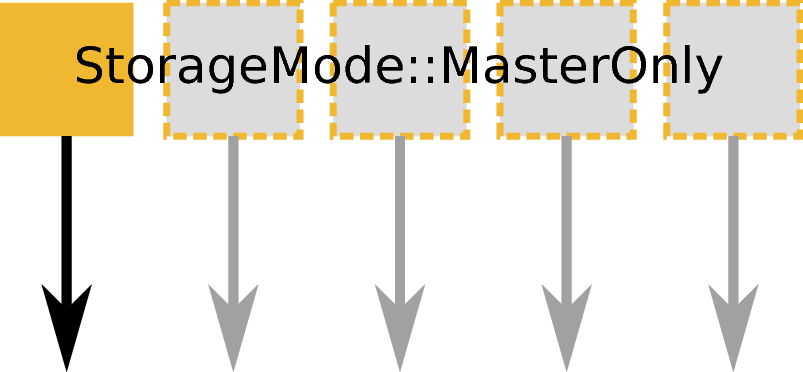
ExecutionMode::MasterOnly based on an input workspace with StorageMode::MasterOnly an no output. Example: Save or any other save algorithm.
Just like the storage mode describes how data is stored, and execution mode describes how an algorithm is executed on this data. There are five execution modes (in namespace Parallel):
The use of the word ‘typically’ above is intentional and indicates that there may be other cases. In particular, an algorithm may cause a transition from one storage mode to another, or may take inputs with different storage modes. Examples are given in the series of figures on the right.
To build Mantid with MPI support as described in this document run cmake with the additional option -DMPI_EXPERIMENTAL=ON. This requires boost-mpi and a working MPI installation.
In principle Python scripts that use only algorithms that support MPI can be run with MPI without changes. For example:
from mantid.simpleapi import *
dataX = [1,2,3,4,2,3,4,5,3,4,5,6,4,5,6,7]
dataY = [1,1,1,1,1,1,1,1,1,1,1,1]
dataE = [1,1,1,1,1,1,1,1,1,1,1,1]
# CreateWorkspace has a new property called ParallelStorageMode that allows setting the
# desired storage mode. It defaults to "Parallel::StorageMode::Cloned".
dataWS = CreateWorkspace(DataX=dataX, DataY=dataY, DataE=dataE, NSpec=4, UnitX="Wavelength", ParallelStorageMode="Parallel::StorageMode::Distributed")
ws = Rebin(dataWS, "1,1,7");
print("Histograms: " + str(ws.getNumberHistograms()))
for i in range(ws.getNumberHistograms()):
print("(Local) workspace index: " + str(i))
print(ws.readX(i))
print(ws.readY(i))
Run Python with mpirun and the desired number of MPI ranks:
mpirun -n 3 python test.py
Note that directly using the Mantid Python wrapper mantidpython is not possible, i.e., mpirun -n 3 mantidpython test.py does not work. Instead the correct paths to Mantid and library preloads should be set manually. Alternatively, a modified version of mantidpython that internally uses mpirun to call python could be created.
Possible output:
CreateWorkspace-[Notice] CreateWorkspace started
CreateWorkspace-[Notice] CreateWorkspace successful, Duration 0.02 seconds
Rebin-[Notice] Rebin started
Rebin-[Notice] Rebin successful, Duration 0.01 seconds
Histograms: 2
(Local) workspace index: 0
[ 1. 2. 3. 4. 5. 6. 7.]
[ 1. 1. 1. 0. 0. 0.]
(Local) workspace index: 1
[ 1. 2. 3. 4. 5. 6. 7.]
[ 0. 0. 0. 1. 1. 1.]
Histograms: 1
(Local) workspace index: 0
[ 1. 2. 3. 4. 5. 6. 7.]
[ 0. 1. 1. 1. 0. 0.]
Histograms: 1
(Local) workspace index: 0
[ 1. 2. 3. 4. 5. 6. 7.]
[ 0. 0. 1. 1. 1. 0.]
Output involving the local number of histograms and local indices is obviously not useful for users and should be avoided (see also the section on workspace indices), this example is merely for illustration.
Note that currently Mantid does not support workspaces without spectra, so running above example with more than four MPI ranks fill fail since there are only four spectra. This is probably not a problem in practice.
With many MPI ranks it is common to get spammed by logging output. Since there is not control of output order for multi-line log messages it also tends to become hard to read since output from different ranks get interleaved.
The current solution to this is a logging offset for all but the master rank. By default an offset of 1 is added, i.e., an error message from any rank but rank 0 will be displayed as a warning. The offset can be adjusted in the Mantid properties file, e.g.,
mpi.loggingOffset=3
The drawback of this approach is that information contained in error or warning messages that are specific to a spectrum, such as a missing detector ID, can be hidden or lost. If that is an issue the logging offset can simply be set to 0.
Only MatrixWorkspace and its subclasses support StorageMode::Distributed. All other workspace types, in particular TableWorkspace and MDWorkspace are restricted to StorageMode::MasterOnly and StorageMode::Cloned.
By default an algorithm does not support MPI and any attempt to execute it in an MPI run will throw an exception. MPI support for an algorithm is implemented by means of a couple of virtual methods in the Algorithm base class:
class Algorithm {
// ...
protected:
virtual void execDistributed();
virtual void execMasterOnly();
virtual void execNonMaster();
virtual Parallel::ExecutionMode getParallelExecutionMode(
const std::map<std::string, Parallel::StorageMode> &storageModes) const;
// ...
};
In general it is not necessary to implement all of these methods. For many algorithms it can be sufficient to implement getParallelExecutionMode. This is often the case if an algorithm has only a single input and a single output and treats all spectra independently. In that case the execution mode can simply be determined from the input workspace as follows:
Parallel::ExecutionMode MyAlg::getParallelExecutionMode(
const std::map<std::string, Parallel::StorageMode> &storageModes) const {
// The map key is the property name. If there is only one input workspace it can usually be ignored.
return Parallel::getCorrespondingExecutionMode(storageModes.begin()->second);
}
Here the helper Parallel::getCorrespondingExecutionMode is used to obtain the ‘natural’ execution mode from a storage mode, i.e., ExecutionMode::Identical for StorageMode::Cloned, ExecutionMode::Distributed for StorageMode::Distributed, and ExecutionMode::MasterOnly for StorageMode::MasterOnly. More complex algorithms may require more complex decision mechanism, e.g., when there is more than one input workspace.
If none of the other virtual methods listed above is implemented, Algorithm will run the normal exec() method on all MPI ranks. The exception are non-master ranks if the execution mode is ExecutionMode::MasterOnly – in that case creating a dummy workspace is attempted. This is discussed in more detail in the subsections below.
Identical execution with execution mode ExecutionMode::Identical is usually done for data with storage mode StorageMode::Cloned. Execution is handled by simply calling Algorithm::exec() on all MPI ranks.
A notable exception that has to be kept in mind are algorithms that are saving workspaces or write to other resources, since the file names will be in conflict.
Distributed execution is handled by Algorithm::execDistributed(). By default this simply calls Algorithm::exec(). In many cases this may be perfectly fine and more convenient than reimplementing Algorithm::execDistributed().
The following example illustrates the difference. We can either check for the number of MPI ranks in the normal exec() method:
void MyAlg::exec() {
//// Algorithm logics, e.g., a sum over all spectra ////
if (communicator.size() > 1) {
//// MPI calls, e.g., a global sum ////
}
}
Alternatively, we can implement Algorithm::execDistributed():
void MyAlg::exec() {
//// Algorithm logics ////
}
void MyAlg::execDistributed() {
//// Algorithm logics but in a very different way ////
}
Many algorithms in Mantid will require very little modification for MPI support and thus the first option is likely to be the first choice.
Master-only execution is handled by Algorithm::execMasterOnly(). By default this simply calls Algorithm::exec() on rank 0 and Algorithm::execNonMaster() on all other ranks.
To support running existing Python scripts without significant modification, and to be able to automatically determine execution modes based on input workspaces, workspaces with storage mode StorageMode::MasterOnly also exist on the non-master ranks. The default implementation of Algorithm::execNonMaster() creates an uninitialized (in the case of MatrixWorkspace) workspace of the same type as the input workspace. If Algorithm::execNonMaster() is overridden, any workspaces that are created shall also be uninitialized and should have storage mode StorageMode::MasterOnly.
Given that the workspace on non-master ranks are not initialized, no methods of the workspace should be called, apart from Workspace::storageMode(). Validators on the non-master ranks are thus also disabled.
A typical implementation could look as follows:
void MyAlg::execNonMaster() {
setProperty("OutputWorkspace", Kernel::make_unique<Workspace2D>(
Parallel::StorageMode::MasterOnly));
}
Setting spectrum numbers via the legacy interface MatrixWorkspace::getSpectrum(size_t)::setSpectrumNo(specnum_t) is not supported in MPI runs and will throw an exception. The reason is that spectrum numbers are used to globally identify a spectrum and thus changing a spectrum number must be done globally, i.e., on all MPI ranks. Spectrum numbers should be set by using Indexing::IndexInfo and MatrixWorkspace::setIndexInfo(), or rather by passing the IndexInfo to one of the workspace factory functions from DataObjects/WorkspaceCreation.h.
If a workspace is distributed, i.e., has storage mode StorageMode::Distributed workspaces indices lose their meaning. In particular, MatrixWorkspace::getNumberHistograms() will return the local number of spectra and not the global size of the workspace. For purposes of interaction with the user interface and for internal consistency a global equivalent of the ‘workspace index’ concept has been introduced. This index is represented by Indexing::GlobalSpectrumIndex. [2]
The consequences are as follows:
As described above, the full set of detectors is held on each MPI rank. Thus, algorithms that modify detectors must do so in an identical manner on all MPI ranks. That is, if for example detector positions would be modified in an Algorithm it is not sufficient to do so for all detectors that have a corresponding spectrum on the MPI rank. Instead such a modification must be done for all detectors.
The details of this depend on what exactly an algorithm is supposed to do and a generic recipe cannot be given here. It is however essential to think of this when providing MPI support for an algorithm.
Running the Mantid GUI with MPI support, such as a client GUI with a MPI-based backend, is currently not possible. If it cannot be avoided to add an MPI-related property to an algorithm is shall be made invisible in the GUI. This can be done by adjusting the property settings when implementing Algorithm::init():
#include "MantidKernel/InvisibleProperty.h"
void MyAlg::init() {
// ...
setPropertySettings("MyProperty", Kernel::make_unique<InvisibleProperty>());
}
For unit testing the MPI support of an algorithm a fake backend that can be run without MPI is provided. No modifications to the code under test a required. In the unit test case ParallelRunner from MantidTestHelpers is used to run the algorithm (or other code) under test as if it were part of on MPI run. A typical example could look as follows:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 | #include "MantidTestHelpers/ParallelAlgorithmCreation.h"
#include "MantidTestHelpers/ParallelRunner.h"
namespace {
void run_algorithm(const Parallel::Communicator &comm,
const MyType1 &arbitrary, const MyType2 &arguments) {
// Creating the algorithm with this helper function is the recommended way,
// otherwise the communicator has to be set by hand and the name of the
// output workspace must be set to a different value depending on comm.rank()
// to avoid clashes, since the threading backend in ParallelRunner shares the
// ADS for all 'ranks'.
auto alg = ParallelTestHelpers::create<Mantid::Algorithms::MyAlg>(comm);
alg->setProperty("InputWorkspace", boost::make_shared<WorkspaceTester>());
alg->execute();
Workspace_const_sptr ws = alg->getProperty("OutputWorkspace");
TS_ASSERT_EQUALS(ws->storageMode(), Parallel::StorageMode::Distributed);
}
class MyAlgTest : public CxxTest::TestSuite {
public:
// ...
void test_parallel() {
// Runs run_algorithm in multiple threads. The first argument passed to
// run_algorithm is of type Parallel::Communicator and is guaranteed to
// have size() > 1, i.e., more than one rank, in at least one call to
// run_algorithm (it is in addition also called with a single 'rank').
ParallelTestHelpers::runParallel(run_algorithm, 42, 42.0);
}
};
|
Here MantidTestHelpers/ParallelAlgorithmCreation.h provides the algorithm factory method ParallelTestHelpers::create<WorkspaceType>. MantidTestHelpers/ParallelRunner.h provides ParallelTestHelpers::runParallel, which uses ParallelRunner with a reasonable default choice for the number of ranks.
When adding MPI support for an algorithm, add it to the table at the end of this document. Potential limitations must be described in the comments.
| Algorithm | Supported modes | Comments |
|---|---|---|
| CreateWorkspace | all | |
| Rebin | all | min and max bin boundaries must be given explicitly |
Footnotes
| [1] | The complexity and overhead of splitting the instrument, in particular given the overhead ensuing from handling all cases exemplified above, led to the decision split only the neutron data based on spectra, but not detectors. |
| [2] | Some will argue that this should be GlobalWorkspaceIndex. However it is not an index of a workspace so the term GlobalSpectrumIndex has been chosen for clarity. On the user interface side this will still be named ‘workspace index’, dropping the ‘global’ since the distinction between global and local indices is irrelevant for users. |
Category: Development