Table of Contents
Create a MDEventWorkspace with events in reciprocal space (Qx, Qy, Qz, Energy) from a SQW file.
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| Filename | Input | string | Mandatory | File of type SQW format. Allowed extensions: [‘.sqw’] |
| OutputWorkspace | Output | MDEventWorkspace | Mandatory | Output IMDEventWorkspace reflecting SQW data read-in. |
| MetadataOnly | Input | boolean | False | Load Metadata without events. |
| OutputFilename | Input | string | If the input SQW file is too large to fit in memory, specify an output NXS file. The MDEventWorkspace will be create with this file as its back-end. Allowed extensions: [‘.nxs’] |
The algorithm takes every pixel defined in the SQW horace file and converts it into an event.
Only top level binning information is currently taken from DND/Image
data. All DND image information is currently ignored and the resulting
MDEvent workspace is in the units of 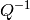 (SQW dimensions,
recalculated to the Crystal? frame, without HKL transformation).
(SQW dimensions,
recalculated to the Crystal? frame, without HKL transformation).
U matrix is set to unity but the B-matrix is read from the SQW and attached to the workspace which may confuse the algorithms which work with MDEventWorkspace produced by Mantid algorithms.
Sqw objects comes in two flavours:
The former has the four main fields fully filled. The fields description is given below. The latter has:
The dnd-type is created from d0d, d1d,... objects for most algorithms so as to use private methods of sqw objects.
DND-type object can not be currently read or understood by Mantid.
String started by:
% -- ignored
* -- used
Main_header:
% main_headerfilename Name of sqw file that is being read, excluding path
% main_headerfilepath Path to sqw file that is being read, including terminating file separator
% main_headertitle Title of sqw data structure
% main_headernfiles Number of spe files that contribute to the sqw object
Header: (scalar structure, or cellarray of scalar structures if more than one spe file)
% header{i}.filename Name of sqw file excluding path
% header{i}.filepath Path to sqw file including terminating file separator
% header{i}.efix Fixed energy (ei or ef depending on emode)
% header{i}.emode Emode=1 direct geometry, =2 indirect geometry, =0 if diffraction ''' Only emode 1 have ever been tried '''
% header{i}.alatt Lattice parameters (Angstroms)
% header{i}.angdeg Lattice angles (deg)
% header{i}.cu First vector defining scattering plane (r.l.u.)
% header{i}.cv Second vector defining scattering plane (r.l.u.)
% header{i}.psi Orientation angle (rad)
% header{i}.omega --|
% header{i}.dpsi | Crystal misorientation description (rad)
% header{i}.gl | (See notes elsewhere e.g. Tobyfit manual
% header{i}.gs --|
% header{i}.en Energy bin boundaries (meV) in the input spe file [column vector]
% header{i}.uoffset Offset of origin of pixel projection axes in r.l.u. and energy i.e. [h; k; l; en] [column vector]
% header{i}.u_to_rlu Matrix (4x4) of pixel projection axes in hkle representation
% u(:,1) first vector - u(1:3,1) r.l.u., u(4,1) energy etc.
% header{i}.ulen Length of pixel projection axes vectors in Ang^-1 or meV [row vector]
% header{i}.ulabel Labels of the pixel projection axes [1x4 cell array of character strings]
The pixel projection axes, u1, u2, u3 are the orthonormal vectors of the crystal Cartesian coordinate frame i.e. u1 || a*, u2 in plane of a*, and b*, and u3 || a* x b*. They form the coordinate frame in which the pixel coordinates are stored in data.pix. The pixel projection axes must necessarily be identical for all contributing spe files.
\ **Detpar**\ :
% detpar.filename Name of file excluding path
% detpar.filepath Path to file including terminating file separator
% detpar.group Row vector of detector group number
% detpar.x2 Row vector of secondary flightpaths (m)
% detpar.phi Row vector of scattering angles (deg)
% detpar.azim Row vector of azimuthal angles (deg)
% (West bank=0 deg, North bank=90 deg etc.)
% detpar.width Row vector of detector widths (m)
% detpar.height Row vector of detector heights (m)
\ **Data**\ :
% data.filename Name of sqw file that is being read, excluding path
% data.filepath Path to sqw file that is being read, including terminating file separator
% data.title Title of sqw data structure
* data.alatt Lattice parameters for data field (Ang^-1)
* data.angdeg Lattice angles for data field (degrees)
% data.uoffset Offset of origin of projection axes in r.l.u. and energy ie. [h; k; l; en] [column vector]
% data.u_to_rlu Matrix (4x4) of projection axes in hkle representation
% u(:,1) first vector - u(1:3,1) r.l.u., u(4,1) energy etc.
% data.ulen Length of projection axes vectors in Ang^-1 or meV [row vector]
% data.ulabel Labels of the projection axes [1x4 cell array of character strings]
% data.iax Index of integration axes into the projection axes [row vector]
% Always in increasing numerical order
% e.g. if data is 2D, data.iax=[1,3] means summation has been performed along u1 and u3 axes
% data.iint Integration range along each of the integration axes. [iint(2,length(iax))]
% e.g. in 2D case above, is the matrix vector [u1_lo, u3_lo; u1_hi, u3_hi]
% data.pax Index of plot axes into the projection axes [row vector]
% Always in increasing numerical order
% e.g. if data is 3D, data.pax=[1,2,4] means u1, u2, u4 axes are x,y,z in any plotting
% 2D, data.pax=[2,4] " u2, u4, axes are x,y in any plotting
% data.p Call array containing bin boundaries along the plot axes [column vectors]
% i.e. row cell array {data.p{1}, data.p{2} ...} (for as many axes as length of data.pax)
% data.dax Index into data.pax of the axes for display purposes. For example we may have
% data.pax=[1,3,4] and data.dax=[3,1,2] This means that the first display axis is data.pax(3)=4,
% the second is data.pax(1)=1, the third is data.pax(2)=3. The reason for data.dax is to allow
% the display axes to be permuted but without the contents of the fields p, s,..pix needing to
% be reordered [row vector]
-----> Large data fields, data for MD image
% data.s Cumulative signal. [size(data.s)=(length(data.p1)-1, length(data.p2)-1, ...)]
% data.e Cumulative variance [size(data.e)=(length(data.p1)-1, length(data.p2)-1, ...)]
% data.npix No. contributing pixels to each bin of the plot axes.
% [size(data.pix)=(length(data.p1)-1, length(data.p2)-1, ...)]
----->
* data.urange True range of the data along each axis [urange(2,4)]
----> Pixels or events data
* data.pix Array containing data for each pixel:
* If npixtot=sum(npix), then pix(9,npixtot) contains:
* u1 -|
* u2 | Coordinates of pixel in the pixel projection axes
* u3 |
* u4 -|
* irun Run index in the header block from which pixel came
* idet Detector group number in the detector listing for the pixel
* ien Energy bin number for the pixel in the array in the (irun)th header
* signal Signal array
* err Error array (variance i.e. error bar squared)
data.s is normalized by the number of pixels, as is the variance data.e. For those elements where data.npix==0, data.s=0 and data.e=0
Parts of the code were written with the idea of generalising functionality at a later stage. However, we can now assume that:
Note
To run these usage examples please first download the usage data, and add these to your path. In MantidPlot this is done using Manage User Directories.
Example - Load 4D workspace form sample SQW file.
# Delete existing workspace if it is already in Mantid as LoadSQW does not support overwriting
# existig workspaces by design
if 'mdws' in mtd:
DeleteWorkspace('mdws')
#
# Load sample sqw file, present in Mantid unit tests as MD workspace
mdws = LoadSQW('test_horace_reader.sqw');
# Check results
print "Workspace type is: ",mdws.id()
print "Workspace has:{0:2} dimensions and contains: {1:4} MD events".format(mdws.getNumDims(),mdws.getNEvents())
Output:
Workspace type is: MDEventWorkspace<MDEvent,4>
Workspace has: 4 dimensions and contains: 580 MD events
Categories: Algorithms | DataHandling | MDAlgorithms