Table of Contents
| Name | Direction | Type | Default | Description |
|---|---|---|---|---|
| LHSWorkspace | Input | MatrixWorkspace | Mandatory | LHS input workspace. |
| RHSWorkspace | Input | MatrixWorkspace | Mandatory | RHS input workspace, must be same type as LHSWorkspace (histogram or point data). |
| OutputWorkspace | Output | MatrixWorkspace | Mandatory | Output stitched workspace. |
| StartOverlap | Input | number | Optional | Start overlap x-value in units of x-axis. |
| EndOverlap | Input | number | Optional | End overlap x-value in units of x-axis. |
| Params | Input | dbl list | Rebinning Parameters. See Rebin for format. If only a single value is provided, start and end are taken from input workspaces. | |
| ScaleRHSWorkspace | Input | boolean | True | Scaling either with respect to LHS workspace or RHS workspace |
| UseManualScaleFactor | Input | boolean | False | True to use a provided value for the scale factor. |
| ManualScaleFactor | Input | number | 1 | Provided value for the scale factor. |
| OutScaleFactor | Output | number | The actual used value for the scaling factor. |
Stitches single histogram Matrix Workspaces together outputting a stitched Matrix Workspace. The type of the input workspaces (histogram or point data) determines the stitch procedure. The x-error values Dx will always be ignored in case of histogram workspaces. Point data workspaces must be consistent, i.e. must have Dx defined or not.
Either the right-hand-side or left-hand-side workspace can be chosen to be scaled. Users can optionally provide Rebin v1 Params, otherwise they are calculated from the input workspaces. Likewise, StartOverlap and EndOverlap are optional. If not provided, then these are taken to be the region of X-axis intersection.
The algorithm workflow for histograms is as follows:
Below is a flowchart illustrating the steps in the algorithm (it assumes ScaleRHSWorkspace is true). Figure on the left corresponds to the workflow when no scale factor is provided, while figure on the right corresponds to workflow with a manual scale factor specified by the user.
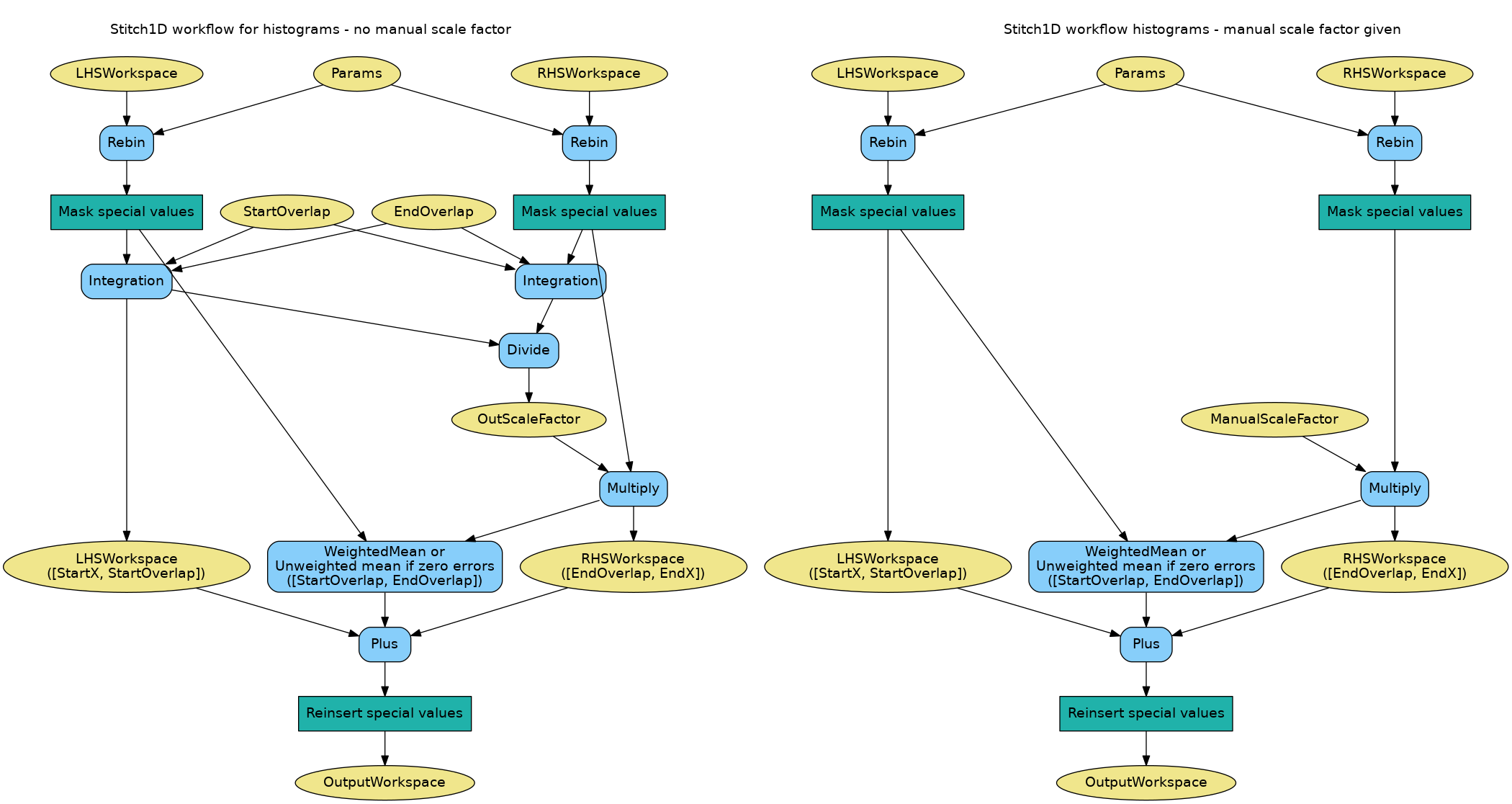
The algorithm workflow for point data is as follows:
Errors are are handled and propagated in every step according to Error Propagation. This includes every child algorithm: Rebin v1, Integration v1, Divide v1, Multiply v1 and WeightedMean v1. In particular, when the scale factor is calculated as the quotient of the left-hand-side integral and the right-hand-side integral, the result is a number with an error associated, and therefore the multiplication of the right-hand-side workspace by this number takes into account its error.
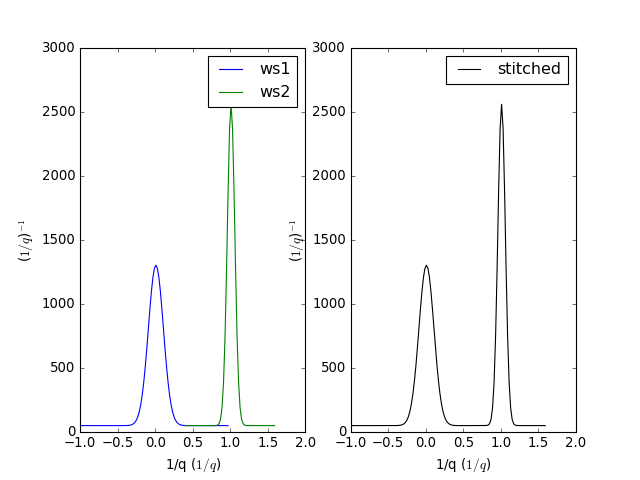
Example - a basic example using stitch1D to stitch two histogram workspaces together.
from mantid.simpleapi import *
import matplotlib.pyplot as plt
import numpy as np
def gaussian(x, mu, sigma):
"""Creates a Gaussian peak centered on mu and with width sigma."""
return (1/ sigma * np.sqrt(2 * np.pi)) * np.exp( - (x-mu)**2 / (2*sigma**2))
# create two histograms with a single peak in each one
x1 = np.arange(-1, 1, 0.02)
x2 = np.arange(0.4, 1.6, 0.02)
ws1 = CreateWorkspace(UnitX="1/q", DataX=x1, DataY=gaussian(x1[:-1], 0, 0.1)+1)
ws2 = CreateWorkspace(UnitX="1/q", DataX=x2, DataY=gaussian(x2[:-1], 1, 0.05)+1)
# stitch the histograms together
stitched, scale = Stitch1D(LHSWorkspace=ws1, RHSWorkspace=ws2, StartOverlap=0.4, EndOverlap=0.6, Params=0.02)
# plot the individual workspaces alongside the stitched one
fig, axs = plt.subplots(nrows=1, ncols=2, subplot_kw={'projection':'mantid'})
axs[0].plot(mtd['ws1'], wkspIndex=0, label='ws1')
axs[0].plot(mtd['ws2'], wkspIndex=0, label='ws2')
axs[0].legend()
axs[1].plot(mtd['stitched'], wkspIndex=0, color='k', label='stitched')
axs[1].legend()
# uncomment the following line to show the plot window
#fig.show()
(Source code, png, hires.png, pdf)
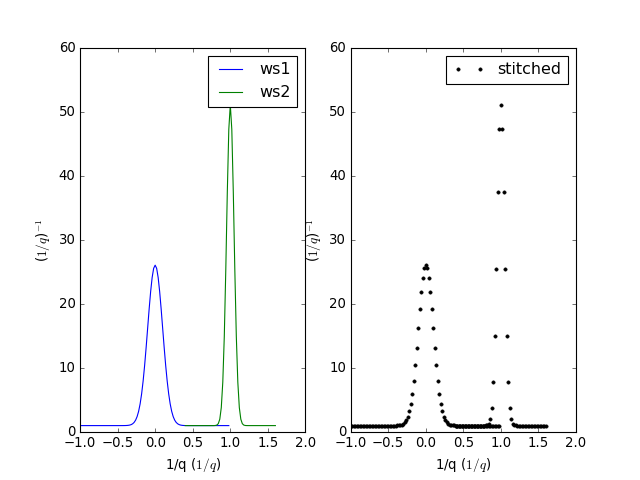
Example - a basic example using stitch1D to stitch two point data workspaces together.
from mantid.simpleapi import *
import matplotlib.pyplot as plt
import numpy as np
def gaussian(x, mu, sigma):
"""Creates a Gaussian peak centered on mu and with width sigma."""
return (1/ sigma * np.sqrt(2 * np.pi)) * np.exp( - (x-mu)**2 / (2*sigma**2))
# create two histograms with a single peak in each one
x1 = np.arange(-1, 1, 0.02)
x2 = np.arange(0.4, 1.6, 0.02)
ws1 = CreateWorkspace(UnitX="1/q", DataX=x1, DataY=gaussian(x1, 0, 0.1)+1)
ws2 = CreateWorkspace(UnitX="1/q", DataX=x2, DataY=gaussian(x2, 1, 0.05)+1)
# stitch the histograms together
stitched, scale = Stitch1D(LHSWorkspace=ws1, RHSWorkspace=ws2, StartOverlap=0.4, EndOverlap=0.6)
# plot the individual workspaces alongside the stitched one
fig, axs = plt.subplots(nrows=1, ncols=2, subplot_kw={'projection':'mantid'})
axs[0].plot(mtd['ws1'], wkspIndex=0, label='ws1')
axs[0].plot(mtd['ws2'], wkspIndex=0, label='ws2')
axs[0].legend()
axs[1].plot(mtd['stitched'], wkspIndex=0, color='k', marker='.', ls='', label='stitched')
axs[1].legend()
# uncomment the following line to show the plot window
#fig.show()
(Source code, png, hires.png, pdf)
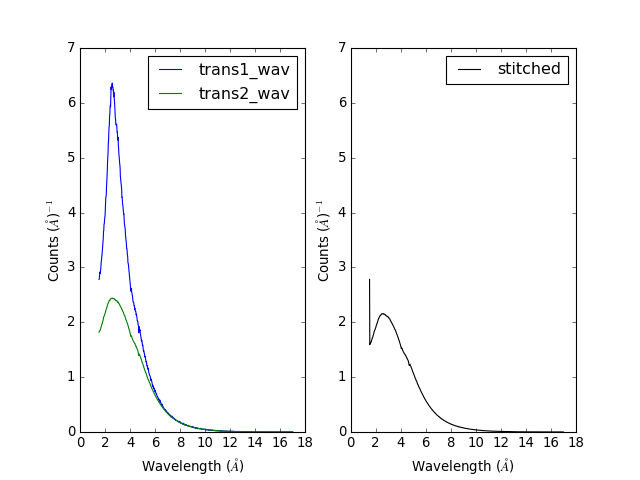
Example - a practical example using reflectometry data and a scale factor.
from mantid.simpleapi import *
import matplotlib.pyplot as plt
try:
trans1 = Load('INTER00013463')
trans2 = Load('INTER00013464')
trans1_wav = CreateTransmissionWorkspaceAuto(trans1)
trans2_wav = CreateTransmissionWorkspaceAuto(trans2)
stitched_wav, y = Stitch1D(trans1_wav, trans2_wav, UseManualScaleFactor=True, ManualScaleFactor=0.85)
# plot the individual and stitched workspaces next to each other
fig, axs = plt.subplots(nrows=1, ncols=2, subplot_kw={'projection':'mantid'})
axs[0].plot(trans1_wav, wkspIndex=0, label=str(trans1_wav))
axs[0].plot(trans2_wav, wkspIndex=0, label=str(trans2_wav))
axs[0].legend()
# use same y scale on both plots
ylimits = axs[0].get_ylim()
axs[1].plot(stitched_wav, wkspIndex=0, color='k', label='stitched')
axs[1].legend()
axs[1].set_ylim(ylimits)
# uncomment the following line to show the plot window
#fig.show()
except ValueError:
print("Cannot load data")
(Source code, png, hires.png, pdf)
Categories: AlgorithmIndex | Reflectometry
C++ source: Stitch1D.cpp (last modified: 2019-07-17)
C++ header: Stitch1D.h (last modified: 2018-10-05)